Stimulation of methane generation from nonproductive coal by addition of nutrients or a microbial consortium
- PMID: 20817801
- PMCID: PMC2976240
- DOI: 10.1128/AEM.00728-10
Stimulation of methane generation from nonproductive coal by addition of nutrients or a microbial consortium
Abstract
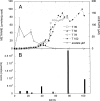
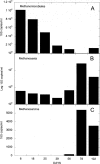
Biogenic formation of methane from coal is of great interest as an underexploited source of clean energy. The goal of some coal bed producers is to extend coal bed methane productivity and to utilize hydrocarbon wastes such as coal slurry to generate new methane. However, the process and factors controlling the process, and thus ways to stimulate it, are poorly understood. Subbituminous coal from a nonproductive well in south Texas was stimulated to produce methane in microcosms when the native population was supplemented with nutrients (biostimulation) or when nutrients and a consortium of bacteria and methanogens enriched from wetland sediment were added (bioaugmentation). The native population enriched by nutrient addition included Pseudomonas spp., Veillonellaceae, and Methanosarcina barkeri. The bioaugmented microcosm generated methane more rapidly and to a higher concentration than the biostimulated microcosm. Dissolved organics, including long-chain fatty acids, single-ring aromatics, and long-chain alkanes accumulated in the first 39 days of the bioaugmented microcosm and were then degraded, accompanied by generation of methane. The bioaugmented microcosm was dominated by Geobacter sp., and most of the methane generation was associated with growth of Methanosaeta concilii. The ability of the bioaugmentation culture to produce methane from coal intermediates was confirmed in incubations of culture with representative organic compounds. This study indicates that methane production could be stimulated at the nonproductive field site and that low microbial biomass may be limiting in situ methane generation. In addition, the microcosm study suggests that the pathway for generating methane from coal involves complex microbial partnerships.
Figures




Similar articles
-
Long-term succession in a coal seam microbiome during in situ biostimulation of coalbed-methane generation.ISME J. 2019 Mar;13(3):632-650. doi: 10.1038/s41396-018-0296-5. Epub 2018 Oct 15. ISME J. 2019. PMID: 30323265 Free PMC article.
-
Cultivation-independent analysis of archaeal and bacterial communities of the formation water in an Indian coal bed to enhance biotransformation of coal into methane.Appl Microbiol Biotechnol. 2012 Feb;93(3):1337-50. doi: 10.1007/s00253-011-3778-1. Epub 2011 Dec 28. Appl Microbiol Biotechnol. 2012. PMID: 22202965
-
Who eats what? Unravelling microbial conversion of coal to methane.FEMS Microbiol Ecol. 2019 Jul 1;95(7):fiz093. doi: 10.1093/femsec/fiz093. FEMS Microbiol Ecol. 2019. PMID: 31216572
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
-
Microbial methanogenesis in subsurface oil and coal.Res Microbiol. 2013 Nov;164(9):959-72. doi: 10.1016/j.resmic.2013.07.004. Epub 2013 Jul 19. Res Microbiol. 2013. PMID: 23872511 Review.
Cited by
-
Subsurface hydrocarbon degradation strategies in low- and high-sulfate coal seam communities identified with activity-based metagenomics.NPJ Biofilms Microbiomes. 2022 Feb 17;8(1):7. doi: 10.1038/s41522-022-00267-2. NPJ Biofilms Microbiomes. 2022. PMID: 35177633 Free PMC article.
-
Methanogenic archaea in subsurface coal seams are biogeographically distinct: an analysis of metagenomically-derived mcrA sequences.Environ Microbiol. 2022 Sep;24(9):4065-4078. doi: 10.1111/1462-2920.16014. Epub 2022 May 10. Environ Microbiol. 2022. PMID: 35437913 Free PMC article.
-
Activity-based, genome-resolved metagenomics uncovers key populations and pathways involved in subsurface conversions of coal to methane.ISME J. 2022 Apr;16(4):915-926. doi: 10.1038/s41396-021-01139-x. Epub 2021 Oct 23. ISME J. 2022. PMID: 34689183 Free PMC article.
-
Microbial methane formation in deep aquifers of a coal-bearing sedimentary basin, Germany.Front Microbiol. 2015 Mar 20;6:200. doi: 10.3389/fmicb.2015.00200. eCollection 2015. Front Microbiol. 2015. PMID: 25852663 Free PMC article.
-
Algal amendment enhances biogenic methane production from coals of different thermal maturity.Front Microbiol. 2023 Mar 10;14:1097500. doi: 10.3389/fmicb.2023.1097500. eCollection 2023. Front Microbiol. 2023. PMID: 36970672 Free PMC article.
References
-
- Anderson, R. T., and D. R. Lovley. 2000. Hexadecane decay by methanogenesis. Nature 404:722-723. - PubMed
-
- Anderson, R. T., J. N. Rooney-Varga, C. V. Gaw, and D. R. Lovley. 1998. Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum-contaminated aquifers. Environ. Sci. Technol. 32:1222-1229.
-
- Bakermans, C., and E. L. Madsen. 2002. Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. - PubMed
-
- Breland, F. C., Jr. 2004. Coalbed methane potential in Louisiana, p. 27-35. In P. D. Warwick (ed.), Selected presentations on coal-bed gas in the eastern United States. U.S. Geological Survey Open-File Report 2004-1273. U.S. Geological Survey, Reston, VA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

