Gene expression profiles in a rabbit model of systemic lupus erythematosus autoantibody production
- PMID: 20817871
- PMCID: PMC2949067
- DOI: 10.4049/jimmunol.1001254
Gene expression profiles in a rabbit model of systemic lupus erythematosus autoantibody production
Abstract
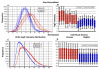
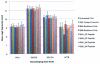
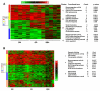
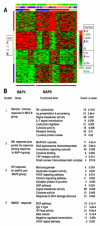
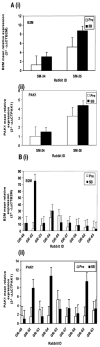
We previously reported the establishment of a rabbit (Oryctolagus cuniculus) model in which peptide immunization led to production of lupus-like autoantibodies including anti-Sm, -RNP, -SS-A, -SS-B, and -dsDNA characteristic of those produced in systemic lupus erythematosus (SLE) patients. Some neurologic symptoms in the form of seizures and nystagmus were observed. The animals used in the previous and in the current study were from a National Institute of Allergy and Infectious Diseases colony of rabbits that were pedigreed, Ig-allotype defined, but not inbred. Their genetic heterogeneity may correspond to that found among patients of a given ethnicity. We extended the information about this rabbit model by microarray-based expression profiling. We first demonstrated that human expression arrays could be used with rabbit RNA to yield information on molecular pathways. We then designed a study evaluating gene expression profiles in eight groups of control and treated rabbits (47 rabbits in total). Genes significantly upregulated in treated rabbits were associated with NK cytotoxicity, Ag presentation, leukocyte migration, cytokine activity, protein kinases, RNA spliceosomal ribonucleoproteins, intracellular signaling cascades, and glutamate receptor activity. These results link increased immune activation with upregulation of components associated with neurologic and anti-RNP responses, demonstrating the utility of the rabbit model to uncover biological pathways related to SLE-induced clinical symptoms, including neuropsychiatric lupus. Our finding of distinct gene expression patterns in rabbits that made anti-dsDNA compared with those that only made other anti-nuclear Abs should be further investigated in subsets of SLE patients with different autoantibody profiles.
Figures
References
-
- Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl. Res. 2009;153:4–10. - PubMed
-
- Jönsen A, Bengtsson AA, Nived O, Truedsson L, Sturfelt G. Gene-environment interactions in the aetiology of systemic lupus erythematosus. Autoimmunity. 2007;40:613–617. - PubMed
-
- Cohen-Solal JF, Jeganathan V, Hill L, Kawabata D, Rodriguez-Pinto D, Grimaldi C, Diamond B. Hormonal regulation of B-cell function and systemic lupus erythematosus. Lupus. 2008;17:528–532. - PubMed
-
- Werth VP. Cutaneous lupus: insights into pathogenesis and disease classification. Bull. NYU Hosp. Jt. Dis. 2007;65:200–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

