Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro
- PMID: 20817910
- PMCID: PMC2978605
- DOI: 10.1074/jbc.M110.169003
Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro
Abstract
Initial attachment of bacteriophage P22 to the Salmonella host cell is known to be mediated by interactions between lipopolysaccharide (LPS) and the phage tailspike proteins (TSP), but the events that subsequently lead to DNA injection into the bacterium are unknown. We used the binding of a fluorescent dye and DNA accessibility to DNase and restriction enzymes to analyze DNA ejection from phage particles in vitro. Ejection was specifically triggered by aggregates of purified Salmonella LPS but not by LPS with different O-antigen structure, by lipid A, phospholipids, or soluble O-antigen polysaccharide. This suggests that P22 does not use a secondary receptor at the bacterial outer membrane surface. Using phage particles reconstituted with purified mutant TSP in vitro, we found that the endorhamnosidase activity of TSP degrading the O-antigen polysaccharide was required prior to DNA ejection in vitro and DNA replication in vivo. If, however, LPS was pre-digested with soluble TSP, it was no longer able to trigger DNA ejection, even though it still contained five O-antigen oligosaccharide repeats. Together with known data on the structure of LPS and phage P22, our results suggest a molecular model. In this model, tailspikes position the phage particles on the outer membrane surface for DNA ejection. They force gp26, the central needle and plug protein of the phage tail machine, through the core oligosaccharide layer and into the hydrophobic portion of the outer membrane, leading to refolding of the gp26 lazo-domain, release of the plug, and ejection of DNA and pilot proteins.
Figures


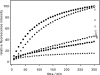
 , P22tWT control without LPS. The standard deviation of fluorescence signals between repeated experiments was below 0.5%.
, P22tWT control without LPS. The standard deviation of fluorescence signals between repeated experiments was below 0.5%.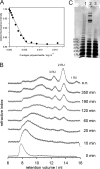

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

