Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex
- PMID: 20817927
- PMCID: PMC3017601
- DOI: 10.1093/nar/gkq749
Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex
Abstract
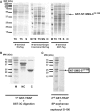
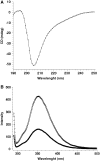
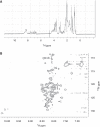
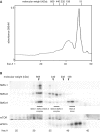
SMG-9 is part of a protein kinase complex, SMG1C, which consists of the SMG-1 kinase, SMG-8 and SMG-9. SMG1C mediated phosphorylation of Upf1 triggers nonsense-mediated mRNA decay (NMD), a eukaryotic surveillance pathway that detects and targets for degradation mRNAs harboring premature translation termination codons. Here, we have characterized SMG-9, showing that it comprises an N-terminal 180 residue intrinsically disordered region (IDR) followed by a well-folded C-terminal domain. Both domains are required for SMG-1 binding and the integrity of the SMG1C complex, whereas the C-terminus is sufficient to interact with SMG-8. In addition, we have found that SMG-9 assembles in vivo into SMG-9:SMG-9 and, most likely, SMG-8:SMG-9 complexes that are not constituents of SMG1C. SMG-9 self-association is driven by interactions between the C-terminal domains and surprisingly, some SMG-9 oligomers are completely devoid of SMG-1 and SMG-8. We propose that SMG-9 has biological functions beyond SMG1C, as part of distinct SMG-9-containing complexes. Some of these complexes may function as intermediates potentially regulating SMG1C assembly, tuning the activity of SMG-1 with the NMD machinery. The structural malleability of IDRs could facilitate the transit of SMG-9 through several macromolecular complexes.
Figures


References
-
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. - PubMed
-
- Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009;16:107–113. - PubMed
-
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

