Liver cytochrome P450 3A ubiquitination in vivo by gp78/autocrine motility factor receptor and C terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligases: physiological and pharmacological relevance
- PMID: 20819951
- PMCID: PMC2975210
- DOI: 10.1074/jbc.M110.167189
Liver cytochrome P450 3A ubiquitination in vivo by gp78/autocrine motility factor receptor and C terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligases: physiological and pharmacological relevance
Abstract
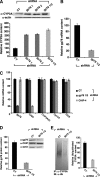
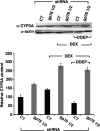
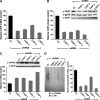
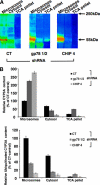
CYP3A4 is a dominant human liver cytochrome P450 enzyme engaged in the metabolism and disposition of >50% of clinically relevant drugs and held responsible for many adverse drug-drug interactions. CYP3A4 and its mammalian liver CYP3A orthologs are endoplasmic reticulum (ER)-anchored monotopic proteins that undergo ubiquitin (Ub)-dependent proteasomal degradation (UPD) in an ER-associated degradation (ERAD) process. These integral ER proteins are ubiquitinated in vivo, and in vitro studies have identified the ER-integral gp78 and the cytosolic co-chaperone, CHIP (C terminus of Hsp70-interacting protein), as the relevant E3 Ub-ligases, along with their cognate E2 Ub-conjugating enzymes UBC7 and UbcH5a, respectively. Using lentiviral shRNA templates targeted against each of these Ub-ligases, we now document that both E3s are indeed physiologically involved in CYP3A ERAD/UPD in cultured rat hepatocytes. Accordingly, specific RNAi resulted in ≈80% knockdown of each hepatic Ub-ligase, with a corresponding ≈2.5-fold CYP3A stabilization. Surprisingly, however, such stabilization resulted in increased levels of functionally active CYP3A, thereby challenging the previous notion that E3 recognition and subsequent ERAD of CYP3A proteins required ab initio their structural and/or functional inactivation. Furthermore, coexpression in HepG2 cells of both CYP3A4 and gp78, but not its functionally inactive RING-finger mutant, resulted in enhanced CYP3A4 loss greater than that in corresponding cells expressing only CYP3A4. Stabilization of a functionally active CYP3A after RNAi knockdown of either of the E3s, coupled with the increased CYP3A4 loss on gp78 or CHIP coexpression, suggests that ERAD-associated E3 Ub-ligases can influence clinically relevant drug metabolism by effectively regulating the physiological CYP3A content and consequently its function.
Figures



Similar articles
-
Multisite phosphorylation of human liver cytochrome P450 3A4 enhances Its gp78- and CHIP-mediated ubiquitination: a pivotal role of its Ser-478 residue in the gp78-catalyzed reaction.Mol Cell Proteomics. 2012 Feb;11(2):M111.010132. doi: 10.1074/mcp.M111.010132. Epub 2011 Nov 17. Mol Cell Proteomics. 2012. PMID: 22101235 Free PMC article.
-
Human liver cytochrome P450 3A4 ubiquitination: molecular recognition by UBC7-gp78 autocrine motility factor receptor and UbcH5a-CHIP-Hsc70-Hsp40 E2-E3 ubiquitin ligase complexes.J Biol Chem. 2015 Feb 6;290(6):3308-32. doi: 10.1074/jbc.M114.611525. Epub 2014 Dec 1. J Biol Chem. 2015. PMID: 25451919 Free PMC article.
-
CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases.Arch Biochem Biophys. 2009 Mar 1;483(1):66-74. doi: 10.1016/j.abb.2008.12.001. Epub 2008 Dec 10. Arch Biochem Biophys. 2009. PMID: 19103148 Free PMC article.
-
CHIP and gp78-mediated ubiquitination of CYP3A4: Implications for the pharmacology of anticancer agents.Cancer Biol Ther. 2011 Mar 15;11(6):549-51. doi: 10.4161/cbt.11.6.14834. Epub 2011 Mar 15. Cancer Biol Ther. 2011. PMID: 21270532 Free PMC article. Review.
-
Hepatic cytochromes P450: structural degrons and barcodes, posttranslational modifications and cellular adapters in the ERAD-endgame.Drug Metab Rev. 2016 Aug;48(3):405-33. doi: 10.1080/03602532.2016.1195403. Epub 2016 Jun 20. Drug Metab Rev. 2016. PMID: 27320797 Free PMC article. Review.
Cited by
-
Membrane Protein Quantity Control at the Endoplasmic Reticulum.J Membr Biol. 2017 Aug;250(4):379-392. doi: 10.1007/s00232-016-9931-0. Epub 2016 Oct 14. J Membr Biol. 2017. PMID: 27743014 Free PMC article. Review.
-
Regulation of cytochrome P450 enzyme activity and expression by nitric oxide in the context of inflammatory disease.Drug Metab Rev. 2020 Nov;52(4):455-471. doi: 10.1080/03602532.2020.1817061. Epub 2020 Sep 8. Drug Metab Rev. 2020. PMID: 32898444 Free PMC article. Review.
-
Ubiquitin-dependent proteasomal degradation of human liver cytochrome P450 2E1: identification of sites targeted for phosphorylation and ubiquitination.J Biol Chem. 2011 Mar 18;286(11):9443-56. doi: 10.1074/jbc.M110.176685. Epub 2011 Jan 5. J Biol Chem. 2011. PMID: 21209460 Free PMC article.
-
Hepatic cytochrome P450 ubiquitination: conformational phosphodegrons for E2/E3 recognition?IUBMB Life. 2014 Feb;66(2):78-88. doi: 10.1002/iub.1247. Epub 2014 Feb 3. IUBMB Life. 2014. PMID: 24488826 Free PMC article. Review.
-
Liver cytochrome P450 3A endoplasmic reticulum-associated degradation: a major role for the p97 AAA ATPase in cytochrome P450 3A extraction into the cytosol.J Biol Chem. 2011 Feb 4;286(5):3815-28. doi: 10.1074/jbc.M110.186981. Epub 2010 Nov 24. J Biol Chem. 2011. PMID: 21107009 Free PMC article.
References
-
- Guengerich F. P. (2005) in Cytochrome P450: Structure, Mechanism and Biochemistry (Ortiz de Montellano P. ed) pp. 377–530, Kluwer-Academic/Plenum Press, New York
-
- Correia M. A., Decker C., Sugiyama K., Caldera P., Bornheim L., Wrighton S. A., Rettie A. E., Trager W. F. (1987) Arch. Biochem. Biophys. 258, 436–451 - PubMed
-
- Correia M. A., Davoll S. H., Wrighton S. A., Thomas P. E. (1992) Arch. Biochem. Biophys. 297, 228–238 - PubMed
-
- Wang H. F., Figueiredo Pereira M. E., Correia M. A. (1999) Arch. Biochem. Biophys. 365, 45–53 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

