Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme
- PMID: 20822536
- PMCID: PMC3092116
- DOI: 10.1186/gm186
Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme
Abstract
Background: Coordinated efforts to collect large-scale data sets provide a basis for systems level understanding of complex diseases. In order to translate these fragmented and heterogeneous data sets into knowledge and medical benefits, advanced computational methods for data analysis, integration and visualization are needed.
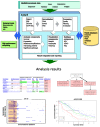
Methods: We introduce a novel data integration framework, Anduril, for translating fragmented large-scale data into testable predictions. The Anduril framework allows rapid integration of heterogeneous data with state-of-the-art computational methods and existing knowledge in bio-databases. Anduril automatically generates thorough summary reports and a website that shows the most relevant features of each gene at a glance, allows sorting of data based on different parameters, and provides direct links to more detailed data on genes, transcripts or genomic regions. Anduril is open-source; all methods and documentation are freely available.
Results: We have integrated multidimensional molecular and clinical data from 338 subjects having glioblastoma multiforme, one of the deadliest and most poorly understood cancers, using Anduril. The central objective of our approach is to identify genetic loci and genes that have significant survival effect. Our results suggest several novel genetic alterations linked to glioblastoma multiforme progression and, more specifically, reveal Moesin as a novel glioblastoma multiforme-associated gene that has a strong survival effect and whose depletion in vitro significantly inhibited cell proliferation. All analysis results are available as a comprehensive website.
Conclusions: Our results demonstrate that integrated analysis and visualization of multidimensional and heterogeneous data by Anduril enables drawing conclusions on functional consequences of large-scale molecular data. Many of the identified genetic loci and genes having significant survival effect have not been reported earlier in the context of glioblastoma multiforme. Thus, in addition to generally applicable novel methodology, our results provide several glioblastoma multiforme candidate genes for further studies.Anduril is available at http://csbi.ltdk.helsinki.fi/anduril/The glioblastoma multiforme analysis results are available at http://csbi.ltdk.helsinki.fi/anduril/tcga-gbm/
Figures

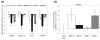
References
LinkOut - more resources
Full Text Sources
Research Materials

