Dynamic deterministic effects propagation networks: learning signalling pathways from longitudinal protein array data
- PMID: 20823327
- PMCID: PMC2935402
- DOI: 10.1093/bioinformatics/btq385
Dynamic deterministic effects propagation networks: learning signalling pathways from longitudinal protein array data
Abstract
Motivation: Network modelling in systems biology has become an important tool to study molecular interactions in cancer research, because understanding the interplay of proteins is necessary for developing novel drugs and therapies. De novo reconstruction of signalling pathways from data allows to unravel interactions between proteins and make qualitative statements on possible aberrations of the cellular regulatory program. We present a new method for reconstructing signalling networks from time course experiments after external perturbation and show an application of the method to data measuring abundance of phosphorylated proteins in a human breast cancer cell line, generated on reverse phase protein arrays.
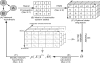
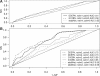
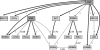
Results: Signalling dynamics is modelled using active and passive states for each protein at each timepoint. A fixed signal propagation scheme generates a set of possible state transitions on a discrete timescale for a given network hypothesis, reducing the number of theoretically reachable states. A likelihood score is proposed, describing the probability of measurements given the states of the proteins over time. The optimal sequence of state transitions is found via a hidden Markov model and network structure search is performed using a genetic algorithm that optimizes the overall likelihood of a population of candidate networks. Our method shows increased performance compared with two different dynamical Bayesian network approaches. For our real data, we were able to find several known signalling cascades from the ERBB signalling pathway.
Availability: Dynamic deterministic effects propagation networks is implemented in the R programming language and available at http://www.dkfz.de/mga2/ddepn/.
Figures


References
-
- Akutsu T, et al. Identification of genetic networks from a small number of gene expression patterns under the Boolean network model. Pac. Symp. Biocomput. 1999:17–28. - PubMed
-
- Cohen P, et al. PDK1, one of the missing links in insulin signal transduction? FEBS Lett. 1997;410:3–10. - PubMed
-
- Durbin R, et al. Biological Sequence Analysis. 1. Cambridge: Cambridge University Press; 1998.
-
- Friedman N, et al. Using Bayesian networks to analyze expression data. J. Comput. Biol. 2000;7:601–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

