Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures
- PMID: 20824106
- PMCID: PMC2932511
- DOI: 10.1128/mBio.00189-10
Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures
Abstract
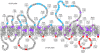
Biosynthesis of B-band lipopolysaccharide (LPS) in Pseudomonas aeruginosa follows the Wzy-dependent pathway, requiring the integral inner membrane proteins Wzx (O-antigen [O-Ag] flippase), Wzy (O-Ag polymerase), and WaaL (O-Ag ligase). For an important first step in deciphering the mechanisms of LPS assembly, we set out to map the membrane topology of these proteins. Random and targeted 3'wzx, wzy, and waaL truncations were fused to a phoA-lacZalpha dual reporter capable of displaying both alkaline phosphatase and beta-galactosidase activity. The results from truncation fusion expression and the corresponding differential enzyme activity ratios allowed for the assignment of specific regions of the proteins to cytoplasmic, transmembrane (TM), or periplasmic loci. Protein orientation in the inner membrane was confirmed via C-terminal fusion to green fluorescent protein. Our data revealed unique TM domain properties in these proteins, particularly for Wzx, indicating the potential for a charged pore. Novel periplasmic and cytoplasmic loop domains were also uncovered, with the latter in Wzy and WaaL revealing tracts consistent with potential Walker A/B motifs.
Figures



Similar articles
-
The WaaL O-antigen lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases.Glycobiology. 2012 Feb;22(2):288-99. doi: 10.1093/glycob/cwr150. Epub 2011 Oct 7. Glycobiology. 2012. PMID: 21983211
-
Escherichia coli and Pseudomonas aeruginosa lipopolysaccharide O-antigen ligases share similar membrane topology and biochemical properties.Mol Microbiol. 2018 Oct;110(1):95-113. doi: 10.1111/mmi.14085. Epub 2018 Oct 3. Mol Microbiol. 2018. PMID: 30047569
-
Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy).Mol Microbiol. 1998 Jun;28(6):1211-22. doi: 10.1046/j.1365-2958.1998.00884.x. Mol Microbiol. 1998. PMID: 9680210
-
Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway.Can J Microbiol. 2014 Nov;60(11):697-716. doi: 10.1139/cjm-2014-0595. Epub 2014 Sep 16. Can J Microbiol. 2014. PMID: 25358682 Review.
-
Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria.Environ Microbiol. 2013 Apr;15(4):1001-15. doi: 10.1111/j.1462-2920.2012.02890.x. Epub 2012 Sep 27. Environ Microbiol. 2013. PMID: 23016929 Review.
Cited by
-
The Polymerization of Aeromonas hydrophila AH-3 O-Antigen LPS: Concerted Action of WecP and Wzy.PLoS One. 2015 Jul 10;10(7):e0131905. doi: 10.1371/journal.pone.0131905. eCollection 2015. PLoS One. 2015. PMID: 26161781 Free PMC article.
-
Progress in Our Understanding of Wzx Flippase for Translocation of Bacterial Membrane Lipid-Linked Oligosaccharide.J Bacteriol. 2017 Dec 5;200(1):e00154-17. doi: 10.1128/JB.00154-17. Print 2018 Jan 1. J Bacteriol. 2017. PMID: 28696276 Free PMC article. Review.
-
Recent insights into Wzy polymerases and lipopolysaccharide O-antigen biosynthesis.J Bacteriol. 2025 Apr 17;207(4):e0041724. doi: 10.1128/jb.00417-24. Epub 2025 Mar 11. J Bacteriol. 2025. PMID: 40066993 Free PMC article. Review.
-
Recent advances in understanding of enterobacterial common antigen synthesis and regulation.Open Biol. 2025 Jan;15(7):250055. doi: 10.1098/rsob.250055. Epub 2025 Jul 2. Open Biol. 2025. PMID: 40592470 Free PMC article. Review.
-
Inhibition of Cronobacter sakazakii Virulence Factors by Citral.Sci Rep. 2017 Feb 24;7:43243. doi: 10.1038/srep43243. Sci Rep. 2017. PMID: 28233814 Free PMC article.
References
-
- Lyczak J. B., Cannon C. L., Pier G. B. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 - PubMed
-
- Schroeder T. H., Reiniger N., Meluleni G., Grout M., Coleman F. T., Pier G. B. 2001. Transgenic cystic fibrosis mice exhibit reduced early clearance of Pseudomonas aeruginosa from the respiratory tract. J. Immunol. 166:7410–7418 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

