Transcriptional reprogramming in nonhuman primate (rhesus macaque) tuberculosis granulomas
- PMID: 20824205
- PMCID: PMC2930844
- DOI: 10.1371/journal.pone.0012266
Transcriptional reprogramming in nonhuman primate (rhesus macaque) tuberculosis granulomas
Abstract
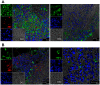
Background: In response to Mtb infection, the host remodels the infection foci into a dense mass of cells known as the granuloma. The key objective of the granuloma is to contain the spread of Mtb into uninfected regions of the lung. However, it appears that Mtb has evolved mechanisms to resist killing in the granuloma. Profiling granuloma transcriptome will identify key immune signaling pathways active during TB infection. Such studies are not possible in human granulomas, due to various confounding factors. Nonhuman Primates (NHPs) infected with Mtb accurately reflect human TB in clinical and pathological contexts.
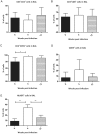
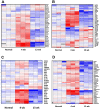
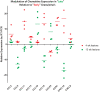
Methodology/principal findings: We studied transcriptomics of granuloma lesions in the lungs of NHPs exhibiting active TB, during early and late stages of infection. Early TB lesions were characterized by a highly pro-inflammatory environment, expressing high levels of immune signaling pathways involving IFNgamma, TNFalpha, JAK, STAT and C-C/C-X-C chemokines. Late TB lesions, while morphologically similar to the early ones, exhibited an overwhelming silencing of the inflammatory response. Reprogramming of the granuloma transcriptome was highly significant. The expression of approximately two-thirds of all genes induced in early lesions was later repressed.
Conclusions/significance: The transcriptional characteristics of TB granulomas undergo drastic changes during the course of infection. The overwhelming reprogramming of the initial pro-inflammatory surge in late lesions may be a host strategy to limit immunopathology. We propose that these host profiles can predict changes in bacterial replication and physiology, perhaps serving as markers for latency and reactivation.
Conflict of interest statement
Figures


References
-
- Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–40. - PubMed
-
- Gandhi NR, Moll A, Sturm AW, Pawinski R, Govendar T, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–80. - PubMed
-
- Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. - PubMed
-
- Orme IM. Prospects for new vaccines against tuberculosis. Trends Microbiol. 1995;3:401–4. - PubMed
-
- Basaraba RJ. Experimental tuberculosis: the role of comparative pathology in the discovery of improved tuberculosis treatment strategies. Tuberculosis. 2008;88(Suppl 1):S35–47. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

