Structural analysis of a highly glycosylated and unliganded gp120-based antigen using mass spectrometry
- PMID: 20825246
- PMCID: PMC2957511
- DOI: 10.1021/bi1011332
Structural analysis of a highly glycosylated and unliganded gp120-based antigen using mass spectrometry
Abstract
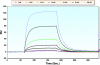
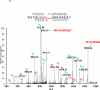
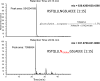
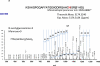
Structural characterization of the HIV-1 envelope protein gp120 is very important for providing an understanding of the protein's immunogenicity and its binding to cell receptors. So far, the crystallographic structure of gp120 with an intact V3 loop (in the absence of a CD4 coreceptor or antibody) has not been determined. The third variable region (V3) of the gp120 is immunodominant and contains glycosylation signatures that are essential for coreceptor binding and entry of the virus into T-cells. In this study, we characterized the structure of the outer domain of gp120 with an intact V3 loop (gp120-OD8) purified from Drosophila S2 cells utilizing mass spectrometry-based approaches. We mapped the glycosylation sites and calculated the glycosylation occupancy of gp120-OD8; 11 sites from 15 glycosylation motifs were determined as having high-mannose or hybrid glycosylation structures. The specific glycan moieties of nine glycosylation sites from eight unique glycopeptides were determined by a combination of ECD and CID MS approaches. Hydroxyl radical-mediated protein footprinting coupled with mass spectrometry analysis was employed to provide detailed information about protein structure of gp120-OD8 by directly identifying accessible and hydroxyl radical-reactive side chain residues. Comparison of gp120-OD8 experimental footprinting data with a homology model derived from the ligated CD4-gp120-OD8 crystal structure revealed a flexible V3 loop structure in which the V3 tip may provide contacts with the rest of the protein while residues in the V3 base remain solvent accessible. In addition, the data illustrate interactions between specific sugar moieties and amino acid side chains potentially important to the gp120-OD8 structure.
Figures




Similar articles
-
Structure of a V3-containing HIV-1 gp120 core.Science. 2005 Nov 11;310(5750):1025-8. doi: 10.1126/science.1118398. Science. 2005. PMID: 16284180 Free PMC article.
-
Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition.Virology. 2003 Sep 1;313(2):387-400. doi: 10.1016/s0042-6822(03)00294-0. Virology. 2003. PMID: 12954207
-
Glycosylation of the core of the HIV-1 envelope subunit protein gp120 is not required for native trimer formation or viral infectivity.J Biol Chem. 2017 Jun 16;292(24):10197-10219. doi: 10.1074/jbc.M117.788919. Epub 2017 Apr 26. J Biol Chem. 2017. PMID: 28446609 Free PMC article.
-
Structural studies of human HIV-1 V3 antibodies.Hum Antibodies. 2005;14(3-4):73-80. Hum Antibodies. 2005. PMID: 16720977 Review.
-
High-Resolution Hydroxyl Radical Protein Footprinting: Biophysics Tool for Drug Discovery.Annu Rev Biophys. 2018 May 20;47:315-333. doi: 10.1146/annurev-biophys-070317-033123. Epub 2018 Mar 14. Annu Rev Biophys. 2018. PMID: 29539273 Review.
Cited by
-
Integrating mass spectrometry of intact protein complexes into structural proteomics.Proteomics. 2012 May;12(10):1547-64. doi: 10.1002/pmic.201100520. Proteomics. 2012. PMID: 22611037 Free PMC article. Review.
-
Mass spectrometry coupled experiments and protein structure modeling methods.Int J Mol Sci. 2013 Oct 15;14(10):20635-57. doi: 10.3390/ijms141020635. Int J Mol Sci. 2013. PMID: 24132151 Free PMC article. Review.
-
Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting.Anal Chem. 2011 Oct 1;83(19):7234-41. doi: 10.1021/ac200567u. Epub 2011 Aug 1. Anal Chem. 2011. PMID: 21770468 Free PMC article.
-
Mapping a Conformational Epitope of Hemagglutinin A Using Native Mass Spectrometry and Ultraviolet Photodissociation.Anal Chem. 2020 Sep 1;92(17):11869-11878. doi: 10.1021/acs.analchem.0c02237. Epub 2020 Aug 12. Anal Chem. 2020. PMID: 32867493 Free PMC article.
-
Eliciting neutralizing antibodies with gp120 outer domain constructs based on M-group consensus sequence.Virology. 2014 Aug;462-463:363-76. doi: 10.1016/j.virol.2014.06.006. Virology. 2014. PMID: 25046154 Free PMC article.
References
-
- Ostermeier C, Iwata S, Ludwig B, Michel H. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat Struct Biol. 1995;2(10):842–846. - PubMed
-
- Kwong PD, Wyatt R, Desjardins E, Robinson J, Culp JS, Hellmig BD, Sweet RW, Sodroski J, Hendrickson WA. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J Biol Chem. 1999;274(7):4115–4123. - PubMed
-
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8(12):1329–1339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

