Asymmetric and non-uniform evolution of recently duplicated human genes
- PMID: 20825637
- PMCID: PMC2942815
- DOI: 10.1186/1745-6150-5-54
Asymmetric and non-uniform evolution of recently duplicated human genes
Abstract
Background: Gene duplications are a source of new genes and protein functions. The innovative role of duplication events makes families of paralogous genes an interesting target for studies in evolutionary biology. Here we study global trends in the evolution of human genes that resulted from recent duplications.
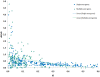
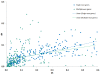
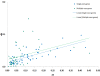
Results: The pressure of negative selection is weaker during a short time immediately after a duplication event. Roughly one fifth of genes in paralogous gene families are evolving asymmetrically: one of the proteins encoded by two closest paralogs accumulates amino acid substitutions significantly faster than its partner. This asymmetry cannot be explained by differences in gene expression levels. In asymmetric gene pairs the number of deleterious mutations is increased in one copy, while decreased in the other copy as compared to genes constituting non-asymmetrically evolving pairs. The asymmetry in the rate of synonymous substitutions is much weaker and not significant.
Conclusions: The increase of negative selection pressure over time after a duplication event seems to be a major trend in the evolution of human paralogous gene families. The observed asymmetry in the evolution of paralogous genes shows that in many cases one of two gene copies remains practically unchanged, while the other accumulates functional mutations. This supports the hypothesis that slowly evolving gene copies preserve their original functions, while fast evolving copies obtain new specificities or functions.
Figures

Similar articles
-
Faster evolving Drosophila paralogs lose expression rate and ubiquity and accumulate more non-synonymous SNPs.Biol Direct. 2014 Jan 17;9:2. doi: 10.1186/1745-6150-9-2. Biol Direct. 2014. PMID: 24438455 Free PMC article.
-
Different evolutionary patterns between young duplicate genes in the human genome.Genome Biol. 2003;4(9):R56. doi: 10.1186/gb-2003-4-9-r56. Epub 2003 Sep 1. Genome Biol. 2003. PMID: 12952535 Free PMC article.
-
Selection in the evolution of gene duplications.Genome Biol. 2002;3(2):RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008. Epub 2002 Jan 14. Genome Biol. 2002. PMID: 11864370 Free PMC article.
-
Role of selection in fixation of gene duplications.J Theor Biol. 2006 Mar 21;239(2):141-51. doi: 10.1016/j.jtbi.2005.08.033. Epub 2005 Oct 20. J Theor Biol. 2006. PMID: 16242725 Review.
-
Gene duplication as a driver of plant morphogenetic evolution.Curr Opin Plant Biol. 2014 Feb;17:43-8. doi: 10.1016/j.pbi.2013.11.002. Epub 2013 Nov 28. Curr Opin Plant Biol. 2014. PMID: 24507493 Review.
Cited by
-
Recurrent sequence evolution after independent gene duplication.BMC Evol Biol. 2020 Aug 8;20(1):98. doi: 10.1186/s12862-020-01660-1. BMC Evol Biol. 2020. PMID: 32770961 Free PMC article.
-
Comparing the Statistical Fate of Paralogous and Orthologous Sequences.Genetics. 2016 Oct;204(2):475-482. doi: 10.1534/genetics.116.193912. Epub 2016 Jul 29. Genetics. 2016. PMID: 27474728 Free PMC article.
-
To the beat of a different drum: determinants implicated in the asymmetric sequence divergence of Caenorhabditis elegans paralogs.BMC Evol Biol. 2013 Mar 27;13:73. doi: 10.1186/1471-2148-13-73. BMC Evol Biol. 2013. PMID: 23530733 Free PMC article.
-
The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013.Nucleic Acids Res. 2013 Jan;41(Database issue):D1063-9. doi: 10.1093/nar/gks1262. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203882 Free PMC article.
-
Long-term asymmetrical acceleration of protein evolution after gene duplication.Genome Biol Evol. 2014 Jul 28;6(8):1949-55. doi: 10.1093/gbe/evu159. Genome Biol Evol. 2014. PMID: 25070510 Free PMC article.
References
-
- Ohta T. Role of gene duplication in evolution. Genome. 1989;31:304–310. - PubMed
-
- Canestro C, Catchen JM, Rodriguez-Mari A, Yokoi H, Postlethwait JH. Consequences of lineage-specific gene loss on functional evolution of surviving paralogs: ALDH1A and retinoic acid signaling in vertebrate genomes. PLoS Genet. 2009;5:e1000496. doi: 10.1371/journal.pgen.1000496. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

