Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling
- PMID: 20826176
- PMCID: PMC3038626
- DOI: 10.1016/j.visres.2010.08.039
Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling
Abstract
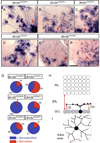
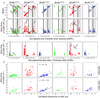
The mammalian retina contains more than 50 distinct neuronal types, which are broadly classified into several major classes: photoreceptor, bipolar, horizontal, amacrine, and ganglion cells. Although some of the developmental mechanisms involved in the differentiation of retinal ganglion cells (RGCs) are beginning to be understood, there is little information regarding the genetic and molecular determinants of the distinct morphologies of the 15-20 mammalian RGC cell types. Previous work has shown that the transcription factor Brn3b/Pou4f2 plays a major role in the development and survival of many RGCs. The roles of the closely related family members, Brn3a/Pou4f1 and Brn3c/Pou4f3 in RGC development are less clear. Using a genetically-directed method for sparse cell labeling and sparse conditional gene ablation in mice, we describe here the sets of RGC types in which each of the three Brn3/Pou4f transcription factors are expressed and the consequences of ablating these factors on the development of RGC morphologies.
Published by Elsevier Ltd.
Figures


References
-
- Amthor FR, Oyster CW, Takahashi ES. Morphology of on-off direction-selective ganglion cells in the rabbit retina. Brain Res. 1984;298(1):187–190. - PubMed
-
- Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. - PubMed
-
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

