Protein tyrosine phosphatase PTPN14 is a regulator of lymphatic function and choanal development in humans
- PMID: 20826270
- PMCID: PMC2933336
- DOI: 10.1016/j.ajhg.2010.08.008
Protein tyrosine phosphatase PTPN14 is a regulator of lymphatic function and choanal development in humans
Abstract
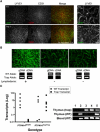
The lymphatic vasculature is essential for the recirculation of extracellular fluid, fat absorption, and immune function and as a route of tumor metastasis. The dissection of molecular mechanisms underlying lymphangiogenesis has been accelerated by the identification of tissue-specific lymphatic endothelial markers and the study of congenital lymphedema syndromes. We report the results of genetic analyses of a kindred inheriting a unique autosomal-recessive lymphedema-choanal atresia syndrome. These studies establish linkage of the trait to chromosome 1q32-q41 and identify a loss-of-function mutation in PTPN14, which encodes a nonreceptor tyrosine phosphatase. The causal role of PTPN14 deficiency was confirmed by the generation of a murine Ptpn14 gene trap model that manifested lymphatic hyperplasia with lymphedema. Biochemical studies revealed a potential interaction between PTPN14 and the vascular endothelial growth factor receptor 3 (VEGFR3), a receptor tyrosine kinase essential for lymphangiogenesis. These results suggest a unique and conserved role for PTPN14 in the regulation of lymphatic development in mammals and a nonconserved role in choanal development in humans.
2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Karpanen T., Alitalo K. Molecular biology and pathology of lymphangiogenesis. Annu. Rev. Pathol. 2008;3:367–397. - PubMed
-
- Tammela T., Alitalo K. Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 2010;140:460–476. - PubMed
-
- Ferrell R.E., Levinson K.L., Esman J.H., Kimak M.A., Lawrence E.C., Barmada M.M., Finegold D.N. Hereditary lymphedema: evidence for linkage and genetic heterogeneity. Hum. Mol. Genet. 1998;7:2073–2078. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

