Stable interaction between the human proliferating cell nuclear antigen loader complex Ctf18-replication factor C (RFC) and DNA polymerase {epsilon} is mediated by the cohesion-specific subunits, Ctf18, Dcc1, and Ctf8
- PMID: 20826785
- PMCID: PMC2966076
- DOI: 10.1074/jbc.M110.166710
Stable interaction between the human proliferating cell nuclear antigen loader complex Ctf18-replication factor C (RFC) and DNA polymerase {epsilon} is mediated by the cohesion-specific subunits, Ctf18, Dcc1, and Ctf8
Abstract
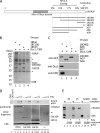
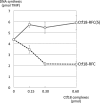
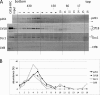
One of the proliferating cell nuclear antigen loader complexes, Ctf18-replication factor C (RFC), is involved in sister chromatid cohesion. To examine its relationship with factors involved in DNA replication, we performed a proteomics analysis of Ctf18-interacting proteins. We found that Ctf18 interacts with a replicative DNA polymerase, DNA polymerase ε (pol ε). Co-immunoprecipitation with recombinant Ctf18-RFC and pol ε demonstrated that their binding is direct and mediated by two distinct interactions, one weak and one stable. Three subunits that are specifically required for cohesion in yeast, Ctf18, Dcc1, and Ctf8, formed a trimeric complex (18-1-8) and together enabled stable binding with pol ε. The C-terminal 23-amino acid stretch of Ctf18 was necessary for the trimeric association of 18-1-8 and was required for the stable interaction. The weak interaction was observed with alternative loader complexes including Ctf18-RFC(5), which lacks Dcc1 and Ctf8, suggesting that the common loader structures, including the RFC small subunits (RFC2-5), are responsible for the weak interaction. The two interaction modes, mediated through distinguishable structures of Ctf18-RFC, both occurred through the N-terminal half of pol ε, which includes the catalytic domain. The addition of Ctf18-RFC or Ctf18-RFC(5) to the DNA synthesis reaction caused partial inhibition and stimulation, respectively. Thus, Ctf18-RFC has multiple interactions with pol ε that promote polymorphic modulation of DNA synthesis. We propose that their interaction alters the DNA synthesis mode to enable the replication fork to cooperate with the establishment of cohesion.
Figures



Similar articles
-
A second proliferating cell nuclear antigen loader complex, Ctf18-replication factor C, stimulates DNA polymerase eta activity.J Biol Chem. 2007 Jul 20;282(29):20906-14. doi: 10.1074/jbc.M610102200. Epub 2007 May 31. J Biol Chem. 2007. PMID: 17545166
-
The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA.Proc Natl Acad Sci U S A. 2003 Sep 2;100(18):10237-42. doi: 10.1073/pnas.1434308100. Epub 2003 Aug 20. Proc Natl Acad Sci U S A. 2003. PMID: 12930902 Free PMC article.
-
Human CTF18-RFC clamp-loader complexed with non-synthesising DNA polymerase ε efficiently loads the PCNA sliding clamp.Nucleic Acids Res. 2017 May 5;45(8):4550-4563. doi: 10.1093/nar/gkx096. Nucleic Acids Res. 2017. PMID: 28199690 Free PMC article.
-
Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication.Adv Exp Med Biol. 2017;1042:135-162. doi: 10.1007/978-981-10-6955-0_7. Adv Exp Med Biol. 2017. PMID: 29357057 Review.
-
The PCNA-RFC families of DNA clamps and clamp loaders.Prog Nucleic Acid Res Mol Biol. 2004;78:227-60. doi: 10.1016/S0079-6603(04)78006-X. Prog Nucleic Acid Res Mol Biol. 2004. PMID: 15210332 Review.
Cited by
-
Sequencing of candidate chromosome instability genes in endometrial cancers reveals somatic mutations in ESCO1, CHTF18, and MRE11A.PLoS One. 2013 Jun 3;8(6):e63313. doi: 10.1371/journal.pone.0063313. Print 2014. PLoS One. 2013. PMID: 23755103 Free PMC article.
-
Chicory: Understanding the Effects and Effectors of This Functional Food.Nutrients. 2022 Feb 23;14(5):957. doi: 10.3390/nu14050957. Nutrients. 2022. PMID: 35267932 Free PMC article.
-
Control of Genome Integrity by RFC Complexes; Conductors of PCNA Loading onto and Unloading from Chromatin during DNA Replication.Genes (Basel). 2017 Jan 26;8(2):52. doi: 10.3390/genes8020052. Genes (Basel). 2017. PMID: 28134787 Free PMC article. Review.
-
A conserved Polϵ binding module in Ctf18-RFC is required for S-phase checkpoint activation downstream of Mec1.Nucleic Acids Res. 2015 Oct 15;43(18):8830-8. doi: 10.1093/nar/gkv799. Epub 2015 Aug 6. Nucleic Acids Res. 2015. PMID: 26250113 Free PMC article.
-
Claspin recruits Cdc7 kinase for initiation of DNA replication in human cells.Nat Commun. 2016 Jul 12;7:12135. doi: 10.1038/ncomms12135. Nat Commun. 2016. PMID: 27401717 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

