UniFrac: an effective distance metric for microbial community comparison
- PMID: 20827291
- PMCID: PMC3105689
- DOI: 10.1038/ismej.2010.133
UniFrac: an effective distance metric for microbial community comparison
Figures
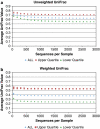
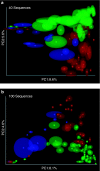
References
-
- Chao A, Chazdon RL, Colwell RK, Shen TJ. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett. 2005;8:148–159.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

