Multivariate Hawkes process models of the occurrence of regulatory elements
- PMID: 20828413
- PMCID: PMC2949889
- DOI: 10.1186/1471-2105-11-456
Multivariate Hawkes process models of the occurrence of regulatory elements
Abstract
Background: A central question in molecular biology is how transcriptional regulatory elements (TREs) act in combination. Recent high-throughput data provide us with the location of multiple regulatory regions for multiple regulators, and thus with the possibility of analyzing the multivariate distribution of the occurrences of these TREs along the genome.
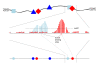
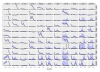
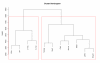
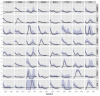
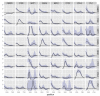
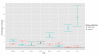
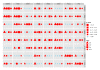
Results: We present a model of TRE occurrences known as the Hawkes process. We illustrate the use of this model by analyzing two different publically available data sets. We are able to model, in detail, how the occurrence of one TRE is affected by the occurrences of others, and we can test a range of natural hypotheses about the dependencies among the TRE occurrences. In contrast to earlier efforts, pre-processing steps such as clustering or binning are not needed, and we thus retain information about the dependencies among the TREs that is otherwise lost. For each of the two data sets we provide two results: first, a qualitative description of the dependencies among the occurrences of the TREs, and second, quantitative results on the favored or avoided distances between the different TREs.
Conclusions: The Hawkes process is a novel way of modeling the joint occurrences of multiple TREs along the genome that is capable of providing new insights into dependencies among elements involved in transcriptional regulation. The method is available as an R package from http://www.math.ku.dk/~richard/ppstat/.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

