Oscillating gene expression determines competence for periodic Arabidopsis root branching
- PMID: 20829477
- PMCID: PMC2976612
- DOI: 10.1126/science.1191937
Oscillating gene expression determines competence for periodic Arabidopsis root branching
Abstract
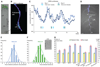
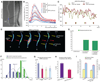
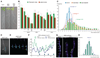
Plants and animals produce modular developmental units in a periodic fashion. In plants, lateral roots form as repeating units along the root primary axis; however, the developmental mechanism regulating this process is unknown. We found that cyclic expression pulses of a reporter gene mark the position of future lateral roots by establishing prebranch sites and that prebranch site production and root bending are periodic. Microarray and promoter-luciferase studies revealed two sets of genes oscillating in opposite phases at the root tip. Genetic studies show that some oscillating transcriptional regulators are required for periodicity in one or both developmental processes. This molecular mechanism has characteristics that resemble molecular clock-driven activities in animal species.
Figures

Comment in
-
Plant science. Oscillating roots.Science. 2010 Sep 10;329(5997):1290-1. doi: 10.1126/science.1195572. Science. 2010. PMID: 20829471 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

