The BioPAX community standard for pathway data sharing
- PMID: 20829833
- PMCID: PMC3001121
- DOI: 10.1038/nbt.1666
The BioPAX community standard for pathway data sharing
Erratum in
- Nat Biotechnol. 2010 Dec;28(12):1308
- Nat Biotechnol. 2012 Apr;30(4):365. Reubenacker, Oliver [corrected to Ruebenacker, Oliver]
Abstract
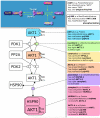
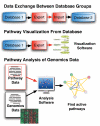
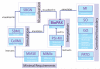
Biological Pathway Exchange (BioPAX) is a standard language to represent biological pathways at the molecular and cellular level and to facilitate the exchange of pathway data. The rapid growth of the volume of pathway data has spurred the development of databases and computational tools to aid interpretation; however, use of these data is hampered by the current fragmentation of pathway information across many databases with incompatible formats. BioPAX, which was created through a community process, solves this problem by making pathway data substantially easier to collect, index, interpret and share. BioPAX can represent metabolic and signaling pathways, molecular and genetic interactions and gene regulation networks. Using BioPAX, millions of interactions, organized into thousands of pathways, from many organisms are available from a growing number of databases. This large amount of pathway data in a computable form will support visualization, analysis and biological discovery.
Figures



References
-
- Nicholson DE. The evolution of the IUBMB-Nicholson maps. IUBMB life. 2000;50:341–344. - PubMed
-
- Demir E, et al. PATIKA: an integrated visual environment for collaborative construction and analysis of cellular pathways. Bioinformatics. 2002;18:996–1003. - PubMed
-
- Fukuda K, Takagi T. Knowledge representation of signal transduction pathways. Bioinformatics. 2001;17:829–837. - PubMed
Publication types
MeSH terms
Grants and funding
- U54 HG004028/HG/NHGRI NIH HHS/United States
- R01 GM071962/GM/NIGMS NIH HHS/United States
- P41 HG004118/HG/NHGRI NIH HHS/United States
- R01GM071962-07/GM/NIGMS NIH HHS/United States
- P41HG004118/HG/NHGRI NIH HHS/United States
- 1R13GM076939/GM/NIGMS NIH HHS/United States
- P30 CA069533/CA/NCI NIH HHS/United States
- R01 GM070923/GM/NIGMS NIH HHS/United States
- U54 AI081680/AI/NIAID NIH HHS/United States
- BB/F010516/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 RR022971/RR/NCRR NIH HHS/United States
- R13 GM076939/GM/NIGMS NIH HHS/United States
- U24 GM077678/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

