Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases
- PMID: 20830216
- PMCID: PMC2935701
- DOI: 10.1021/ja952786k
Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases
Abstract
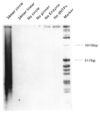
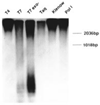
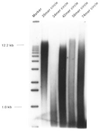
We report that small, single-stranded circular DNA oligonucleotides 26 to 74 nucleotides (nt) in size can behave as catalytic templates for DNA synthesis by several DNA polymerase enzymes. The DNA products are repeating end-to-end multimeric copies of the synthetic circular DNAs, and range from 1 000 to > 12 000 nucleotides in length. Several aspects of this reaction are unusual: first, the synthesis proceeds efficiently despite the curvature and small size of the circles, some of which have diameters significantly smaller than that of the enzyme itself. Second, the synthesis can proceed hundreds of times around the circle, while rolling replication of larger circular plasmid DNAs requires other proteins for processive synthesis. Finally, the synthesis scheme produces multiple copies of the template without the requirement for either heating or cooling cycles and requires less than stoichiometric amounts of primer, unlike other DNA synthesis methods. We report on the scope of this reaction, and demonstrate that the multimeric products can be cleaved enzymatically to short, sequence-defined oligodeoxynucleotides. This new approach to DNA synthesis may be a practical way to produce useful repeating DNAs, and combined with DNA cleavage strategies, it may represent a useful enzymatic approach to short, sequence-defined oligodeoxynucleotides.
Figures







References
-
- Kornberg A. DNA Replication. San Francisco: W. H. Freeman; 1980.
-
- Kool ET. J. Am. Chem. Soc. 1991;113:6265–6266.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
