Use of metallopeptide based mimics demonstrates that the metalloprotein nitrile hydratase requires two oxidized cysteinates for catalytic activity
- PMID: 20831172
- PMCID: PMC3570060
- DOI: 10.1021/ic101765h
Use of metallopeptide based mimics demonstrates that the metalloprotein nitrile hydratase requires two oxidized cysteinates for catalytic activity
Abstract
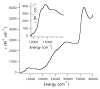
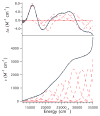
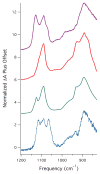
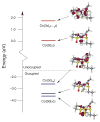
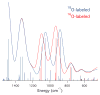
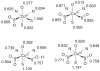
Nitrile hydratases (NHases) are non-heme Fe(III) or non-corrin Co(III) containing metalloenzymes that possess an N(2)S(3) ligand environment with nitrogen donors derived from amidates and sulfur donors derived from cysteinates. A closely related enzyme is thiocyanate hydrolase (SCNase), which possesses a nearly identical active-site coordination environment as CoNHase. These enzymes are redox inactive and perform hydrolytic reactions; SCNase hydrolyzes thiocyanate anions while NHase converts nitriles into amides. Herein an active CoNHase metallopeptide mimic, [Co(III)NHase-m1] (NHase-m1 = AcNH-CCDLP-CGVYD-PA-COOH), that contains Co(III) in a similar N(2)S(3) coordination environment as CoNHase is reported. [Co(III)NHase-m1] was characterized by electrospray ionization-mass spectrometry (ESI-MS), gel-permeation chromatography (GPC), Co K-edge X-ray absorption spectroscopy (Co-S: 2.21 Å; Co-N: 1.93 Å), vibrational, and optical spectroscopies. We find that [Co(III)NHase-m1] will perform the catalytic conversion of acrylonitrile into acrylamide with up to 58 turnovers observed after 18 h at 25 °C (pH 8.0). FTIR data used in concert with calculated vibrational data (mPWPW91/aug-cc-TZVPP) demonstrates that the active form of [Co(III)NHase-m1] has a ligated SO(2) (ν = 1091 cm(-1)) moiety and a ligated protonated SO(H) (ν = 928 cm(-1)) moiety; when only one oxygenated cysteinate ligand (i.e., a mono-SO(2) coordination motif) or the bis-SO(2) coordination motif are found within [Co(III)NHase-m1] no catalytic activity is observed. Calculations of the thermodynamics of ligand exchange (B3LYP/aug-cc-TZVPP) suggest that the reason for this is that the SO(2)/SO(H) equatorial ligand motif promotes both water dissociation from the Co(III)-center and nitrile coordination to the Co(III)-center. In contrast, the under- or overoxidized motifs will either strongly favor a five coordinate Co(III)-center or strongly favor water binding to the Co(III)-center over nitrile binding.
Figures








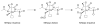
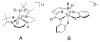

Similar articles
-
Modulation of the pK(a) of metal-bound water via oxidation of thiolato sulfur in model complexes of Co(III) containing nitrile hydratase: insight into possible effect of cysteine oxidation in Co-nitrile hydratase.Inorg Chem. 2003 Sep 8;42(18):5751-61. doi: 10.1021/ic030088s. Inorg Chem. 2003. PMID: 12950226
-
Why is there an "inert" metal center in the active site of nitrile hydratase? Reactivity and ligand dissociation from a five-coordinate Co(III) nitrile hydratase model.J Am Chem Soc. 2001 Jan 24;123(3):463-8. doi: 10.1021/ja002642s. J Am Chem Soc. 2001. PMID: 11456548 Free PMC article.
-
Two arginine residues in the substrate pocket predominantly control the substrate selectivity of thiocyanate hydrolase.J Biosci Bioeng. 2013 Jul;116(1):22-7. doi: 10.1016/j.jbiosc.2013.01.013. Epub 2013 Mar 1. J Biosci Bioeng. 2013. PMID: 23453853
-
Fe(III) and Co(III) centers with carboxamido nitrogen and modified sulfur coordination: lessons learned from nitrile hydratase.Acc Chem Res. 2004 Apr;37(4):253-60. doi: 10.1021/ar0301532. Acc Chem Res. 2004. PMID: 15096062 Review.
-
Nitrile hydratases (NHases): at the interface of academia and industry.Biotechnol Adv. 2010 Nov-Dec;28(6):725-41. doi: 10.1016/j.biotechadv.2010.05.020. Epub 2010 May 25. Biotechnol Adv. 2010. PMID: 20685247 Review.
Cited by
-
pH Dependent Reversible Formation of a Binuclear Ni2 Metal-Center Within a Peptide Scaffold.Inorganics (Basel). 2019 Jul;7(7):90. doi: 10.3390/inorganics7070090. Epub 2019 Jul 16. Inorganics (Basel). 2019. PMID: 38046130 Free PMC article.
-
Multiple States of Nitrile Hydratase from Rhodococcus equi TG328-2: Structural and Mechanistic Insights from Electron Paramagnetic Resonance and Density Functional Theory Studies.Biochemistry. 2017 Jun 20;56(24):3068-3077. doi: 10.1021/acs.biochem.6b00876. Epub 2017 Jun 2. Biochemistry. 2017. PMID: 28520398 Free PMC article.
-
Sulfur oxygenation in biomimetic non-heme iron-thiolate complexes.Dalton Trans. 2012 Aug 28;41(36):10883-99. doi: 10.1039/c2dt30806a. Dalton Trans. 2012. PMID: 22814765 Free PMC article.
-
Construction of a subunit-fusion nitrile hydratase and discovery of an innovative metal ion transfer pattern.Sci Rep. 2016 Jan 12;6:19183. doi: 10.1038/srep19183. Sci Rep. 2016. PMID: 26755342 Free PMC article.
-
Sequential oxidations of thiolates and the cobalt metallocenter in a synthetic metallopeptide: implications for the biosynthesis of nitrile hydratase.Inorg Chem. 2013 May 6;52(9):5236-45. doi: 10.1021/ic400171z. Epub 2013 Apr 15. Inorg Chem. 2013. PMID: 23587023 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

