CD23/FcεRII: molecular multi-tasking
- PMID: 20831712
- PMCID: PMC2990925
- DOI: 10.1111/j.1365-2249.2010.04210.x
CD23/FcεRII: molecular multi-tasking
Abstract
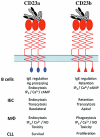
CD23 is the low-affinity receptor for immunoglobulin (Ig)E and plays important roles in the regulation of IgE responses. CD23 can be cleaved from cell surfaces to yield a range of soluble CD23 (sCD23) proteins that have pleiotropic cytokine-like activities. The regions of CD23 responsible for interaction with many of its known ligands, including IgE, CD21, major histocompatibility complex (MHC) class II and integrins, have been identified and help to explain the structure-function relationships within the CD23 protein. Translational studies of CD23 underline its credibility as a target for therapeutic intervention strategies and illustrate its involvement in mediating therapeutic effects of antibodies directed at other targets.
© 2010 The Authors. Clinical and Experimental Immunology © 2010 British Society for Immunology.
Figures



References
-
- Conrad DH, Ford JW, Sturgill JL, Gibb DR. CD23: an overlooked regulator of allergic disease. Curr Allergy Asthma Rep. 2007;7:331–7. - PubMed
-
- Bonnefoy JY, Lecoanet-Henchoz S, Gauchat JF, et al. Structure and functions of CD23. Int Rev Immunol. 1997;16:113–28. - PubMed
-
- Gould HJ, Sutton BJ, Beavil AJ, et al. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003;21:579–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

