Evolutionary evidence of the effect of rare variants on disease etiology
- PMID: 20831747
- PMCID: PMC3652532
- DOI: 10.1111/j.1399-0004.2010.01535.x
Evolutionary evidence of the effect of rare variants on disease etiology
Abstract
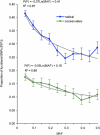
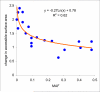
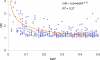
The common disease/common variant hypothesis has been popular for describing the genetic architecture of common human diseases for several years. According to the originally stated hypothesis, one or a few common genetic variants with a large effect size control the risk of common diseases. A growing body of evidence, however, suggests that rare single-nucleotide polymorphisms (SNPs), i.e. those with a minor allele frequency of less than 5%, are also an important component of the genetic architecture of common human diseases. In this study, we analyzed the relevance of rare SNPs to the risk of common diseases from an evolutionary perspective and found that rare SNPs are more likely than common SNPs to be functional and tend to have a stronger effect size than do common SNPs. This observation, and the fact that most of the SNPs in the human genome are rare, suggests that rare SNPs are a crucial element of the genetic architecture of common human diseases. We propose that the next generation of genomic studies should focus on analyzing rare SNPs. Further, targeting patients with a family history of the disease, an extreme phenotype, or early disease onset may facilitate the detection of risk-associated rare SNPs.
© 2010 John Wiley & Sons A/S.
Figures

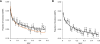
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

