Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia
- PMID: 20831793
- PMCID: PMC2942850
- DOI: 10.1186/1471-2148-10-274
Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia
Abstract
Background: Mitochondrial gene loss and functional transfer to the nucleus is an ongoing process in many lineages of plants, resulting in substantial variation across species in mitochondrial gene content. The Caryophyllaceae represents one lineage that has experienced a particularly high rate of mitochondrial gene loss relative to other angiosperms.
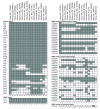
Results: In this study, we report the first complete mitochondrial genome sequence from a member of this family, Silene latifolia. The genome can be mapped as a 253,413 bp circle, but its structure is complicated by a large repeated region that is present in 6 copies. Active recombination among these copies produces a suite of alternative genome configurations that appear to be at or near "recombinational equilibrium". The genome contains the fewest genes of any angiosperm mitochondrial genome sequenced to date, with intact copies of only 25 of the 41 protein genes inferred to be present in the common ancestor of angiosperms. As observed more broadly in angiosperms, ribosomal proteins have been especially prone to gene loss in the S. latifolia lineage. The genome has also experienced a major reduction in tRNA gene content, including loss of functional tRNAs of both native and chloroplast origin. Even assuming expanded wobble-pairing rules, the mitochondrial genome can support translation of only 17 of the 61 sense codons, which code for only 9 of the 20 amino acids. In addition, genes encoding 18S and, especially, 5S rRNA exhibit exceptional sequence divergence relative to other plants. Divergence in one region of 18S rRNA appears to be the result of a gene conversion event, in which recombination with a homologous gene of chloroplast origin led to the complete replacement of a helix in this ribosomal RNA.
Conclusions: These findings suggest a markedly expanded role for nuclear gene products in the translation of mitochondrial genes in S. latifolia and raise the possibility of altered selective constraints operating on the mitochondrial translational apparatus in this lineage.
Figures

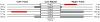





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

