Biosynthetic enzymes of unusual microbial sugars
- PMID: 20832292
- PMCID: PMC4254661
- DOI: 10.1016/j.sbi.2010.08.002
Biosynthetic enzymes of unusual microbial sugars
Abstract
The biological importance of proteins and nucleic acids in the natural world is undeniable, and research efforts on these macromolecules have often overshadowed those directed at carbohydrates. It is now known, however, that carbohydrates not only play roles in energy storage and plant cell wall structure, but are also intimately involved in such processes as fertilization, the immune response, and cell adhesion. Indeed, recent years have seen an explosion in research efforts directed at uncovering and understanding new sugar moieties. The dideoxysugars and trideoxysugars, which are synthesized by a variety of bacteria, fungi, and plants, represent an especially intriguing class of carbohydrates. They are found, for example, on the lipopolysaccharides of some Gram-negative bacteria or on antibacterial agents such as erythromycin. Many of them are formed from simple monosaccharides such as glucose-6-phosphate or fructose-6-phosphate via a myriad of enzymatic reactions including acetylations, aminations, dehydrations, epimerizations, reductions, and methylations. In this review we focus on the recent structural investigations of the bacterial N-acetyltransferases and the PLP-dependent aminotransferases that function on nucleotide-linked sugar substrates.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures

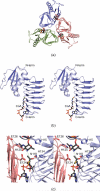

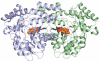
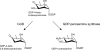
References
-
- Thibodeaux CJ, Melancon CE, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. - PubMed
-
- Raetz CR, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. - PubMed
-
- Olsen LR, Roderick SL. Structure of the Escherichia coli GlmU pyrophosphorylase and acetyltransferase active sites. Biochemistry. 2001;40:1913–1921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

