CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network
- PMID: 20832320
- PMCID: PMC2975053
- DOI: 10.1016/j.tim.2010.07.004
CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network
Abstract
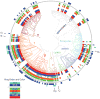
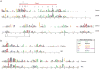
Microbes have chemotactic signaling systems that enable them to detect and follow chemical gradients in their environments. The core of these sensory systems consists of chemoreceptor proteins coupled to the CheA kinase via the scaffold or coupler protein CheW. Some bacterial chemotaxis systems replace or augment CheW with a related protein, CheV, which is less well understood. CheV consists of a CheW domain fused to a receiver domain that is capable of being phosphorylated. Our review of the literature, as well as comparisons of the CheV and CheW sequence and structure, suggest that CheV proteins conserve CheW residues that are crucial for coupling. Phosphorylation of the CheV receiver domain might adjust the efficiency of its coupling and thus allow the system to modulate the response to chemical stimuli in an adaptation process.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


References
-
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. - PubMed
-
- Miller LD, et al. Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol. 2009;66:53–75. - PubMed
-
- Baker MD, et al. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

