The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor
- PMID: 20832723
- PMCID: PMC2950701
- DOI: 10.1016/j.molcel.2010.08.012
The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor
Abstract
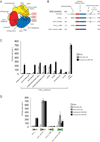
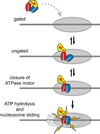
Chromatin remodelers are ATP-driven machines that assemble, slide, and remove nucleosomes from DNA, but how the ATPase motors of remodelers are regulated is poorly understood. Here we show that the double chromodomain unit of the Chd1 remodeler blocks DNA binding and activation of the ATPase motor in the absence of nucleosome substrates. The Chd1 crystal structure reveals that an acidic helix joining the chromodomains can pack against a DNA-binding surface of the ATPase motor. Disruption of the chromodomain-ATPase interface prevents discrimination between nucleosomes and naked DNA and reduces the reliance on the histone H4 tail for nucleosome sliding. We propose that the chromodomains allow Chd1 to distinguish between nucleosomes and naked DNA by physically gating access to the ATPase motor, and we hypothesize that related ATPase motors may employ a similar strategy to discriminate among DNA-containing substrates.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures





References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. - PubMed
-
- Alexiadis V, Lusser A, Kadonaga JT. A conserved N-terminal motif in Rad54 is important for chromatin remodeling and homologous strand pairing. J. Biol. Chem. 2004;279:27824–27829. - PubMed
-
- Beitz E. TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics. 2000;16:135–139. - PubMed
-
- Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007;14:647–652. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

