Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells
- PMID: 20832727
- PMCID: PMC2938080
- DOI: 10.1016/j.molcel.2010.08.013
Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells
Abstract
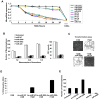
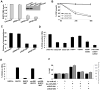
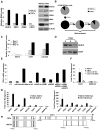
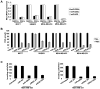
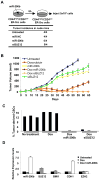
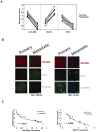
In an inducible oncogenesis model, the miR-200 family is inhibited during CSC formation but not transformation, and inhibition of miR-200b increases CSC formation. Interestingly, miR-200b directly targets Suz12, a subunit of a polycomb repressor complex (PRC2). Loss of miR-200 during CSC formation increases Suz12 expression, Suz12 binding, H3-K27 trimethylation, and Polycomb-mediated repression of the E-cadherin gene. miR-200b expression or Suz12 depletion blocks the formation and maintenance of mammospheres, and in combination with chemotherapy suppresses tumor growth and prolongs remission in mouse xenografts. Conversely, ectopic expression of Suz12 in transformed cells is sufficient to generate CSCs. The miR-200b-Suz12-cadherin pathway is important for CSC growth and invasive ability in genetically distinct breast cancer cells, and its transcriptional signature is observed in metastatic breast tumors. The interaction between miR-200 and Suz12 is highly conserved, suggesting that it represents an ancient regulatory mechanism to control the growth and function of stem cells.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Epigenetic networks and miRNAs in stem cells and cancer.Mol Cell. 2010 Sep 10;39(5):661-3. doi: 10.1016/j.molcel.2010.08.036. Mol Cell. 2010. PMID: 20832717
References
-
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. - PMC - PubMed
-
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

