Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation
- PMID: 20833366
- PMCID: PMC2947937
- DOI: 10.1016/j.devcel.2010.08.012
Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation
Abstract
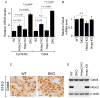
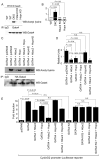
Regulation of chromatin structure via histone modification has recently received intense attention. Here, we demonstrate that the chromatin-modifying enzyme histone deacetylase 2 (Hdac2) functions with a small homeodomain factor, Hopx, to mediate deacetylation of Gata4, which is expressed by cardiac progenitor cells and plays critical roles in the regulation of cardiogenesis. In the absence of Hopx and Hdac2 in mouse embryos, Gata4 hyperacetylation is associated with a marked increase in cardiac myocyte proliferation, upregulation of Gata4 target genes, and perinatal lethality. Hdac2 physically interacts with Gata4, and this interaction is stabilized by Hopx. The ability of Gata4 to transactivate cell cycle genes is impaired by Hopx/Hdac2-mediated deacetylation, and this effect is abrogated by loss of Hdac2-Gata4 interaction. These results suggest that Gata4 is a nonhistone target of Hdac2-mediated deacetylation and that Hdac2, Hopx, and Gata4 coordinately regulate cardiac myocyte proliferation during embryonic development.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

