Mechanisms of control of microRNA biogenesis
- PMID: 20833630
- PMCID: PMC2981492
- DOI: 10.1093/jb/mvq096
Mechanisms of control of microRNA biogenesis
Abstract
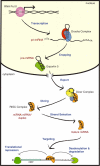
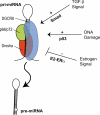
MicroRNAs (miRNAs) are a class of ∼22 nt non-coding RNAs that control diverse biological functions in animals, plants and unicellular eukaryotes by promoting degradation or inhibition of translation of target mRNAs. miRNA expression is often tissue specific and developmentally regulated. Aberrant expression of miRNAs has been linked to developmental abnormalities and human diseases, including cancer and cardiovascular disorders. The recent identification of mechanisms of miRNA biogenesis regulation uncovers that various factors or growth factor signalling pathways control every step of the miRNA biogenesis pathway. Here, we review the mechanisms that control the regulation of miRNA biogenesis discovered in human cells. Further understanding of the mechanisms that control of miRNA biogenesis may allow the development of tools to modulate the expression of specific miRNAs, which is crucial for the development of novel therapies for human disorders derived from aberrant expression of miRNAs.
Figures
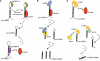
Similar articles
-
Posttranscriptional regulation of microRNA biogenesis in animals.Mol Cell. 2010 May 14;38(3):323-32. doi: 10.1016/j.molcel.2010.03.013. Mol Cell. 2010. PMID: 20471939 Review.
-
A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis.Mol Cell. 2017 Apr 20;66(2):270-284.e13. doi: 10.1016/j.molcel.2017.03.014. Mol Cell. 2017. PMID: 28431233
-
Smad-mediated regulation of microRNA biosynthesis.FEBS Lett. 2012 Jul 4;586(14):1906-12. doi: 10.1016/j.febslet.2012.01.041. Epub 2012 Jan 28. FEBS Lett. 2012. PMID: 22306316 Free PMC article. Review.
-
Diverse small non-coding RNAs in RNA interference pathways.Methods Mol Biol. 2011;764:169-82. doi: 10.1007/978-1-61779-188-8_11. Methods Mol Biol. 2011. PMID: 21748640 Review.
-
Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3.PLoS One. 2011;6(12):e28446. doi: 10.1371/journal.pone.0028446. Epub 2011 Dec 1. PLoS One. 2011. PMID: 22145045 Free PMC article.
Cited by
-
Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers.Int J Mol Sci. 2016 May 23;17(5):791. doi: 10.3390/ijms17050791. Int J Mol Sci. 2016. PMID: 27223281 Free PMC article. Review.
-
Influenza A virus infection of human respiratory cells induces primary microRNA expression.J Biol Chem. 2012 Sep 7;287(37):31027-40. doi: 10.1074/jbc.M112.387670. Epub 2012 Jul 20. J Biol Chem. 2012. PMID: 22822053 Free PMC article.
-
microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles.Am J Physiol Renal Physiol. 2011 Mar;300(3):F602-10. doi: 10.1152/ajprenal.00727.2010. Epub 2011 Jan 12. Am J Physiol Renal Physiol. 2011. PMID: 21228106 Free PMC article. Review.
-
The Role of miRNAs as Key Regulators in the Neoplastic Microenvironment.Mol Biol Int. 2011;2011:839872. doi: 10.4061/2011/839872. Epub 2011 Apr 6. Mol Biol Int. 2011. PMID: 22091413 Free PMC article.
-
Extracellular Vesicles, Cell-Penetrating Peptides and miRNAs as Future Novel Therapeutic Interventions for Parkinson's and Alzheimer's Disease.Biomedicines. 2023 Feb 28;11(3):728. doi: 10.3390/biomedicines11030728. Biomedicines. 2023. PMID: 36979707 Free PMC article. Review.
References
-
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. - PubMed
-
- Lee Y, Ahn C, Han JJ, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
-
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

