Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes
- PMID: 20833811
- PMCID: PMC2953706
- DOI: 10.1128/JB.00624-10
Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes
Abstract
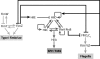
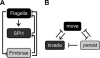
Salmonella enterica, a common food-borne pathogen, differentially regulates the expression of multiple genes during the infection cycle. These genes encode systems related to motility, adhesion, invasion, and intestinal persistence. Key among them is a type three secretion system (T3SS) encoded within Salmonella pathogenicity island 1 (SPI1). In addition to the SPI1 T3SS, other systems, including flagella and type 1 fimbriae, have been implicated in Salmonella pathogenesis. In this study, we investigated the dynamic expression of the flagellar, SPI1, and type 1 fimbrial genes. We demonstrate that these genes are expressed in a temporal hierarchy, beginning with the flagellar genes, followed by the SPI1 genes, and ending with the type 1 fimbrial genes. This hierarchy could mirror the roles of these three systems during the infection cycle. As multiple studies have shown that extensive regulatory cross talk exists between these three systems, we also tested how removing different regulatory links between them affects gene expression dynamics. These results indicate that cross talk is critical for regulating gene expression during transitional phases in the gene expression hierarchy. In addition, we identified a novel regulatory link between flagellar and type 1 fimbrial gene expression dynamics, where we found that the flagellar regulator, FliZ, represses type 1 fimbrial gene expression through the posttranscriptional regulation of FimZ. The significance of these results is that they provide the first systematic study of the effect of regulatory cross talk on the expression dynamics of flagellar, SPI1, and type 1 fimbrial genes.
Figures





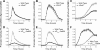
References
-
- Aslanzadeh, J., and L. J. Paulissen. 1992. Role of type 1 and type 3 fimbriae on the adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol. Immunol. 36:351-359. - PubMed
-
- Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

