Semi-supervised recursively partitioned mixture models for identifying cancer subtypes
- PMID: 20834038
- PMCID: PMC2951086
- DOI: 10.1093/bioinformatics/btq470
Semi-supervised recursively partitioned mixture models for identifying cancer subtypes
Abstract
Motivation: Patients with identical cancer diagnoses often progress differently. The disparity we see in disease progression and treatment response can be attributed to the idea that two histologically similar cancers may be completely different diseases on the molecular level. Methods for identifying cancer subtypes associated with patient survival have the capacity to be powerful instruments for understanding the biochemical processes that underlie disease progression as well as providing an initial step toward more personalized therapy for cancer patients. We propose a method called semi-supervised recursively partitioned mixture models (SS-RPMM) that utilizes array-based genetic and patient-level clinical data for finding cancer subtypes that are associated with patient survival.
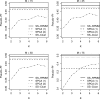
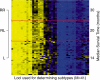
Results: In the proposed SS-RPMM, cancer subtypes are identified using a selected subset of genes that are associated with survival time. Since survival information is used in the gene selection step, this method is semi-supervised. Unlike other semi-supervised clustering classification methods, SS-RPMM does not require specification of the number of cancer subtypes, which is often unknown. In a simulation study, our proposed method compared favorably with other competing semi-supervised methods, including: semi-supervised clustering and supervised principal components analysis. Furthermore, an analysis of mesothelioma cancer data using SS-RPMM, revealed at least two distinct methylation profiles that are informative for survival.
Availability: The analyses implemented in this article were carried out using R (http://www.r.project.org/).
Contact: devin_koestler@brown.edu; e_andres_houseman@brown.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
Similar articles
-
A recursively partitioned mixture model for clustering time-course gene expression data.Transl Cancer Res. 2014;3(3):217-232. doi: 10.3978/j.issn.2218-676X.2014.06.04. Transl Cancer Res. 2014. PMID: 25346887 Free PMC article.
-
Simultaneous gene clustering and subset selection for sample classification via MDL.Bioinformatics. 2003 Jun 12;19(9):1100-9. doi: 10.1093/bioinformatics/btg039. Bioinformatics. 2003. PMID: 12801870
-
Semi-supervised analysis of gene expression profiles for lineage-specific development in the Caenorhabditis elegans embryo.Bioinformatics. 2006 Jul 15;22(14):e417-23. doi: 10.1093/bioinformatics/btl256. Bioinformatics. 2006. PMID: 16873502
-
Semi-supervised methods to predict patient survival from gene expression data.PLoS Biol. 2004 Apr;2(4):E108. doi: 10.1371/journal.pbio.0020108. Epub 2004 Apr 13. PLoS Biol. 2004. PMID: 15094809 Free PMC article.
-
Dissecting cancer heterogeneity--an unsupervised classification approach.Int J Biochem Cell Biol. 2013 Nov;45(11):2574-9. doi: 10.1016/j.biocel.2013.08.014. Epub 2013 Sep 1. Int J Biochem Cell Biol. 2013. PMID: 24004832 Review.
Cited by
-
A recursively partitioned mixture model for clustering time-course gene expression data.Transl Cancer Res. 2014;3(3):217-232. doi: 10.3978/j.issn.2218-676X.2014.06.04. Transl Cancer Res. 2014. PMID: 25346887 Free PMC article.
-
Identification of significant features in DNA microarray data.Wiley Interdiscip Rev Comput Stat. 2013 Jul;5(4):10.1002/wics.1260. doi: 10.1002/wics.1260. Wiley Interdiscip Rev Comput Stat. 2013. PMID: 24244802 Free PMC article.
-
Genome-Scale Methylation Analysis Identifies Immune Profiles and Age Acceleration Associations with Bladder Cancer Outcomes.Cancer Epidemiol Biomarkers Prev. 2023 Oct 2;32(10):1328-1337. doi: 10.1158/1055-9965.EPI-23-0331. Cancer Epidemiol Biomarkers Prev. 2023. PMID: 37527159 Free PMC article.
-
Identification of relevant subtypes via preweighted sparse clustering.Comput Stat Data Anal. 2017 Dec;116:139-154. doi: 10.1016/j.csda.2017.06.003. Epub 2017 Jun 23. Comput Stat Data Anal. 2017. PMID: 29785064 Free PMC article.
-
Comparisons of non-Gaussian statistical models in DNA methylation analysis.Int J Mol Sci. 2014 Jun 16;15(6):10835-54. doi: 10.3390/ijms150610835. Int J Mol Sci. 2014. PMID: 24937687 Free PMC article.
References
-
- Alizadeh AA, et al. Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. - PubMed
-
- Bullinger L, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 2004;350:1605–1616. - PubMed
Publication types
MeSH terms
Grants and funding
- P42ES013660/ES/NIEHS NIH HHS/United States
- P42 ES007373/ES/NIEHS NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- R01CA100679/CA/NCI NIH HHS/United States
- R01CA121147/CA/NCI NIH HHS/United States
- K07CA102327/CA/NCI NIH HHS/United States
- K07 CA102327/CA/NCI NIH HHS/United States
- R01CA126939/CA/NCI NIH HHS/United States
- P01CA134294-01/CA/NCI NIH HHS/United States
- R01 CA100679/CA/NCI NIH HHS/United States
- R01 CA126939/CA/NCI NIH HHS/United States
- P42 ES013660/ES/NIEHS NIH HHS/United States
- R01 CA078609/CA/NCI NIH HHS/United States
- R01 CA121147/CA/NCI NIH HHS/United States
- R01CA078609/CA/NCI NIH HHS/United States
- P42ES007373/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

