Brain fatty acid-binding protein and omega-3/omega-6 fatty acids: mechanistic insight into malignant glioma cell migration
- PMID: 20834042
- PMCID: PMC2978629
- DOI: 10.1074/jbc.M110.170076
Brain fatty acid-binding protein and omega-3/omega-6 fatty acids: mechanistic insight into malignant glioma cell migration
Abstract
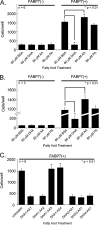
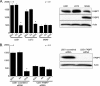
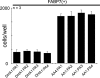
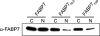
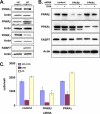
Malignant gliomas (MG) are highly infiltrative tumors that consistently recur despite aggressive treatment. Brain fatty acid-binding protein (FABP7), which binds docosahexaenoic acid (DHA) and arachidonic acid (AA), localizes to sites of tumor infiltration and is associated with a poor prognosis in MG. Manipulation of FABP7 expression in MG cell lines affects cell migration, suggesting a role for FABP7 in tumor infiltration and recurrence. Here, we show that DHA inhibits and AA stimulates migration in an FABP7-dependent manner in U87 MG cells. We demonstrate that DHA binds to and sequesters FABP7 to the nucleus, resulting in decreased cell migration. This anti-migratory effect is partially dependent on peroxisome proliferator-activated receptor γ, a DHA-activated transcription factor. Conversely, AA-bound FABP7 stimulates cell migration by activating cyclooxygenase-2 and reducing peroxisome proliferator-activated receptor γ levels. Our data provide mechanistic insight as to why FABP7 is associated with a poor prognosis in MG and suggest that relative levels of DHA and AA in the tumor environment can make a profound impact on tumor growth properties. We propose that FABP7 and its fatty acid ligands may be key therapeutic targets for controlling the dissemination of MG cells within the brain.
Figures


Similar articles
-
Interaction of brain fatty acid-binding protein with the polyunsaturated fatty acid environment as a potential determinant of poor prognosis in malignant glioma.Prog Lipid Res. 2013 Oct;52(4):562-70. doi: 10.1016/j.plipres.2013.08.004. Epub 2013 Aug 24. Prog Lipid Res. 2013. PMID: 23981365 Free PMC article. Review.
-
FABP7 Facilitates Uptake of Docosahexaenoic Acid in Glioblastoma Neural Stem-like Cells.Nutrients. 2021 Jul 30;13(8):2664. doi: 10.3390/nu13082664. Nutrients. 2021. PMID: 34444824 Free PMC article.
-
The influence of feeding linoleic, gamma-linolenic and docosahexaenoic acid rich oils on rat brain tumor fatty acids composition and fatty acid binding protein 7 mRNA expression.Lipids Health Dis. 2008 Nov 16;7:45. doi: 10.1186/1476-511X-7-45. Lipids Health Dis. 2008. PMID: 19014610 Free PMC article.
-
Nuclear FABP7 immunoreactivity is preferentially expressed in infiltrative glioma and is associated with poor prognosis in EGFR-overexpressing glioblastoma.BMC Cancer. 2006 Apr 19;6:97. doi: 10.1186/1471-2407-6-97. BMC Cancer. 2006. PMID: 16623952 Free PMC article.
-
Transport mechanisms for long-chain polyunsaturated fatty acids in the human placenta.Am J Clin Nutr. 2000 Jan;71(1 Suppl):315S-22S. doi: 10.1093/ajcn/71.1.315s. Am J Clin Nutr. 2000. PMID: 10617989 Review.
Cited by
-
BLBP Is Both a Marker for Poor Prognosis and a Potential Therapeutic Target in Paediatric Ependymoma.Cancers (Basel). 2021 Apr 27;13(9):2100. doi: 10.3390/cancers13092100. Cancers (Basel). 2021. PMID: 33925302 Free PMC article.
-
Neuronal fatty acid-binding protein enhances autophagy and suppresses amyloid-β pathology in a Drosophila model of Alzheimer's disease.PLoS Genet. 2024 Nov 19;20(11):e1011475. doi: 10.1371/journal.pgen.1011475. eCollection 2024 Nov. PLoS Genet. 2024. PMID: 39561115 Free PMC article.
-
Calcineurin regulates nuclear factor I dephosphorylation and activity in malignant glioma cell lines.J Biol Chem. 2013 Aug 16;288(33):24104-15. doi: 10.1074/jbc.M113.455832. Epub 2013 Jul 9. J Biol Chem. 2013. PMID: 23839947 Free PMC article.
-
A Dichotomous Role for FABP7 in Sleep and Alzheimer's Disease Pathogenesis: A Hypothesis.Front Neurosci. 2022 Jun 30;16:798994. doi: 10.3389/fnins.2022.798994. eCollection 2022. Front Neurosci. 2022. PMID: 35844236 Free PMC article.
-
Nuclear FABP7 regulates cell proliferation of wild-type IDH1 glioma through caveolae formation.Mol Oncol. 2022 Jan;16(1):289-306. doi: 10.1002/1878-0261.13130. Epub 2021 Nov 9. Mol Oncol. 2022. PMID: 34716958 Free PMC article.
References
-
- Lin C. L., Lieu A. S., Lee K. S., Yang Y. H., Kuo T. H., Hung M. H., Loh J. K., Yen C. P., Chang C. Z., Howng S. L., Hwang S. L. (2003) Surg. Neurol. 60, 402–406 - PubMed
-
- Godbout R., Bisgrove D. A., Shkolny D., Day R. S., 3rd (1998) Oncogene 16, 1955–1962 - PubMed
-
- Tso C. L., Shintaku P., Chen J., Liu Q., Liu J., Chen Z., Yoshimoto K., Mischel P. S., Cloughesy T. F., Liau L. M., Nelson S. F. (2006) Mol. Cancer Res. 4, 607–619 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

