Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks
- PMID: 20834227
- PMCID: PMC2957216
- DOI: 10.1038/emboj.2010.219
Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks
Abstract
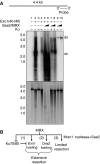
Single-stranded DNA constitutes an important early intermediate for homologous recombination and damage-induced cell cycle checkpoint activation. In Saccharomyces cerevisiae, efficient double-strand break (DSB) end resection requires several enzymes; Mre11/Rad50/Xrs2 (MRX) and Sae2 are implicated in the onset of 5'-strand resection, whereas Sgs1/Top3/Rmi1 with Dna2 and Exo1 are involved in extensive resection. However, the molecular events leading to a switch from the MRX/Sae2-dependent initiation to the Exo1- and Dna2-dependent resection remain unclear. Here, we show that MRX recruits Dna2 nuclease to DSB ends. MRX also stimulates recruitment of Exo1 and antagonizes excess binding of the Ku complex to DSB ends. Using resection assay with purified enzymes in vitro, we found that Ku and MRX regulate the nuclease activity of Exo1 in an opposite way. Efficient loading of Dna2 and Exo1 requires neither Sae2 nor Mre11 nuclease activities. However, Mre11 nuclease activity is essential for resection in the absence of extensive resection enzymes. The results provide new insights into how MRX catalyses end resection and recombination initiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


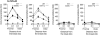



Similar articles
-
Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2.EMBO J. 2010 Oct 6;29(19):3358-69. doi: 10.1038/emboj.2010.193. Epub 2010 Aug 20. EMBO J. 2010. PMID: 20729809 Free PMC article.
-
DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2.Nature. 2010 Sep 2;467(7311):112-6. doi: 10.1038/nature09355. Nature. 2010. PMID: 20811461 Free PMC article.
-
A DNA nick at Ku-blocked double-strand break ends serves as an entry site for exonuclease 1 (Exo1) or Sgs1-Dna2 in long-range DNA end resection.J Biol Chem. 2018 Nov 2;293(44):17061-17069. doi: 10.1074/jbc.RA118.004769. Epub 2018 Sep 17. J Biol Chem. 2018. PMID: 30224356 Free PMC article.
-
Interplay between Sae2 and Rif2 in the regulation of Mre11-Rad50 activities at DNA ends.Curr Opin Genet Dev. 2021 Dec;71:72-77. doi: 10.1016/j.gde.2021.07.001. Epub 2021 Jul 24. Curr Opin Genet Dev. 2021. PMID: 34311383 Review.
-
Structure-function relationships of the Mre11 protein in the control of DNA end bridging and processing.Curr Genet. 2019 Feb;65(1):11-16. doi: 10.1007/s00294-018-0861-5. Epub 2018 Jun 19. Curr Genet. 2019. PMID: 29922906 Review.
Cited by
-
Biochemical and structural characterization of analogs of MRE11 breast cancer-associated mutant F237C.Sci Rep. 2021 Mar 29;11(1):7089. doi: 10.1038/s41598-021-86552-0. Sci Rep. 2021. PMID: 33782469 Free PMC article.
-
14-3-3 proteins restrain the Exo1 nuclease to prevent overresection.J Biol Chem. 2015 May 8;290(19):12300-12. doi: 10.1074/jbc.M115.644005. Epub 2015 Apr 1. J Biol Chem. 2015. PMID: 25833945 Free PMC article.
-
Sae2 controls Mre11 endo- and exonuclease activities by different mechanisms.Nat Commun. 2024 Aug 22;15(1):7221. doi: 10.1038/s41467-024-51493-5. Nat Commun. 2024. PMID: 39174552 Free PMC article.
-
Xrs2 Dependent and Independent Functions of the Mre11-Rad50 Complex.Mol Cell. 2016 Oct 20;64(2):405-415. doi: 10.1016/j.molcel.2016.09.011. Epub 2016 Oct 13. Mol Cell. 2016. PMID: 27746018 Free PMC article.
-
DNA End Resection: Facts and Mechanisms.Genomics Proteomics Bioinformatics. 2016 Jun;14(3):126-130. doi: 10.1016/j.gpb.2016.05.002. Epub 2016 May 27. Genomics Proteomics Bioinformatics. 2016. PMID: 27240470 Free PMC article. Review.
References
-
- Ayyagari R, Gomes XV, Gordenin DA, Burgers PM (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J Biol Chem 278: 1618–1625 - PubMed
-
- Bae SH, Bae KH, Kim JA, Seo YS (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

