Building the developmental oculome: systems biology in vertebrate eye development and disease
- PMID: 20836031
- PMCID: PMC4774529
- DOI: 10.1002/wsbm.59
Building the developmental oculome: systems biology in vertebrate eye development and disease
Abstract
The vertebrate eye is a sophisticated multicomponent organ that has been actively studied for over a century, resulting in the identification of the major embryonic and molecular events involved in its complex developmental program. Data gathered so far provides sufficient information to construct a rudimentary network of the various signaling molecules, transcription factors, and their targets for several key stages of this process. With the advent of genomic technologies, there has been a rapid expansion in our ability to collect and process biological information, and the use of systems-level approaches to study specific aspects of vertebrate eye development has already commenced. This is beginning to result in the definition of the dynamic developmental networks that operate in ocular tissues, and the interactions of such networks between coordinately developing ocular tissues. Such an integrative understanding of the eye by a comprehensive systems-level analysis can be termed the 'oculome', and that of serial developmental stages of the eye as it transits from its initiation to a fully formed functional organ represents the 'developmental oculome'. Construction of the developmental oculome will allow novel mechanistic insights that are essential for organ regeneration-based therapeutic applications, and the generation of computational models for eye disease states to predict the effects of drugs. This review discusses our present understanding of two of the individual components of the developing vertebrate eye--the lens and retina--at both the molecular and systems levels, and outlines the directions and tools required for construction of the developmental oculome.
Figures
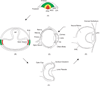
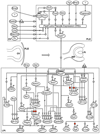


References
-
- McCulloch AD. Modeling the human cardiome in silico. J Nucl Cardiol. 2000;7:496–499. - PubMed
-
- McCulloch AD, Paternostro G. Cardiac systems biology. Ann N Y Acad Sci. 2005;1047:283–295. - PubMed
-
- Srivastava R, Varner J. Emerging technologies: systems biology. Biotechnol Prog. 2007;23:24–27. - PubMed
-
- Monte J, Sakurai H, Bush K, Nigam S. The developmental nephrome: systems biology in the developing kidney. Current Opinion in Nephrology and Hypertension. 2007;16:3–9. - PubMed

