Exploring zebrafish genomic, functional and phenotypic data using ZFIN
- PMID: 20836073
- PMCID: PMC4562798
- DOI: 10.1002/0471250953.bi0118s31
Exploring zebrafish genomic, functional and phenotypic data using ZFIN
Abstract
The zebrafish model organism database (ZFIN) provides a Web resource of zebrafish genomic, genetic, developmental, and phenotypic data. ZFIN curates and integrates data from current literature and from direct data submissions from laboratories. In addition, ZFIN collaborates with other bioinformatics organizations to provide links to other relevant data. These data can be accessed through a variety of Web-based search and display tools. This unit focuses on some of the basic methods to search, visualize, and analyze ZFIN data, including genes, gene expression, mutants, morphants, transgenics, anatomical structures, and antibodies. ZFIN's GBrowse genome viewer, BLAST, and protocol and antibody wikis are also discussed.
Figures























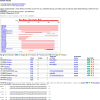
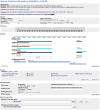







Similar articles
-
ZFIN: enhancements and updates to the Zebrafish Model Organism Database.Nucleic Acids Res. 2011 Jan;39(Database issue):D822-9. doi: 10.1093/nar/gkq1077. Epub 2010 Oct 29. Nucleic Acids Res. 2011. PMID: 21036866 Free PMC article.
-
ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics.Nucleic Acids Res. 2013 Jan;41(Database issue):D854-60. doi: 10.1093/nar/gks938. Epub 2012 Oct 15. Nucleic Acids Res. 2013. PMID: 23074187 Free PMC article.
-
Using ZFIN: Data Types, Organization, and Retrieval.Methods Mol Biol. 2018;1757:307-347. doi: 10.1007/978-1-4939-7737-6_11. Methods Mol Biol. 2018. PMID: 29761463 Free PMC article.
-
Connecting evolutionary morphology to genomics using ontologies: a case study from Cypriniformes including zebrafish.J Exp Zool B Mol Dev Evol. 2007 Sep 15;308(5):655-68. doi: 10.1002/jez.b.21181. J Exp Zool B Mol Dev Evol. 2007. PMID: 17599725 Review.
-
Mitochondrial Disease Sequence Data Resource (MSeqDR): a global grass-roots consortium to facilitate deposition, curation, annotation, and integrated analysis of genomic data for the mitochondrial disease clinical and research communities.Mol Genet Metab. 2015 Mar;114(3):388-96. doi: 10.1016/j.ymgme.2014.11.016. Epub 2014 Dec 4. Mol Genet Metab. 2015. PMID: 25542617 Free PMC article. Review.
Cited by
-
Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens.Front Microbiol. 2015 Feb 4;6:38. doi: 10.3389/fmicb.2015.00038. eCollection 2015. Front Microbiol. 2015. PMID: 25699030 Free PMC article. Review.
-
The mouse Gene Expression Database (GXD): 2011 update.Nucleic Acids Res. 2011 Jan;39(Database issue):D835-41. doi: 10.1093/nar/gkq1132. Epub 2010 Nov 9. Nucleic Acids Res. 2011. PMID: 21062809 Free PMC article.
-
Homozygous splice mutation in CWF19L1 in a Turkish family with recessive ataxia syndrome.Neurology. 2014 Dec 2;83(23):2175-82. doi: 10.1212/WNL.0000000000001053. Epub 2014 Oct 31. Neurology. 2014. PMID: 25361784 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

