New insights into S2P signaling cascades: regulation, variation, and conservation
- PMID: 20836086
- PMCID: PMC3005775
- DOI: 10.1002/pro.496
New insights into S2P signaling cascades: regulation, variation, and conservation
Abstract
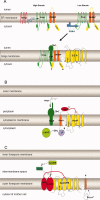
Regulated intramembrane proteolysis (RIP) is a conserved mechanism that regulates signal transduction across the membrane by recruiting membrane-bound proteases to cleave membrane-spanning regulatory proteins. As the first identified protease that performs RIP, the metalloprotease site-2 protease (S2P) has received extensive study during the past decade, and an increasing number of S2P-like proteases have been identified and studied in different organisms; however, some of their substrates and the related S1Ps remain elusive. Here, we review recent research on S2P cascades, including human S2P, E. coli RseP, B. subtilis SpoIVFB and the newly identified S2P homologs. We also discuss the variation and conservation of characterized S2P cascades. The conserved catalytic motif of S2P and prevalence of amino acids of low helical propensity in the transmembrane segments of the substrates suggest a conserved catalytic conformation and mechanism within the S2P family. The review also sheds light on future research on S2P cascades.
Figures



Similar articles
-
Biochemical and structural insights into intramembrane metalloprotease mechanisms.Biochim Biophys Acta. 2013 Dec;1828(12):2873-85. doi: 10.1016/j.bbamem.2013.03.032. Biochim Biophys Acta. 2013. PMID: 24099006 Free PMC article. Review.
-
Site-2 proteases in prokaryotes: regulated intramembrane proteolysis expands to microbial pathogenesis.Microbes Infect. 2006 Jun;8(7):1882-8. doi: 10.1016/j.micinf.2006.02.021. Epub 2006 Apr 27. Microbes Infect. 2006. PMID: 16731018 Review.
-
Involvement of a conserved GFG motif region in substrate binding by RseP, an Escherichia coli S2P protease.Mol Microbiol. 2017 Jun;104(5):737-751. doi: 10.1111/mmi.13659. Epub 2017 Mar 21. Mol Microbiol. 2017. PMID: 28256773
-
The Escherichia coli S2P intramembrane protease RseP regulates ferric citrate uptake by cleaving the sigma factor regulator FecR.J Biol Chem. 2021 Jan-Jun;296:100673. doi: 10.1016/j.jbc.2021.100673. Epub 2021 Apr 16. J Biol Chem. 2021. PMID: 33865858 Free PMC article.
-
Conserved Proline Residues of Bacillus subtilis Intramembrane Metalloprotease SpoIVFB Are Important for Substrate Interaction and Cleavage.J Bacteriol. 2022 Mar 15;204(3):e0038621. doi: 10.1128/JB.00386-21. Epub 2022 Jan 10. J Bacteriol. 2022. PMID: 35007155 Free PMC article.
Cited by
-
Foamy virus envelope protein is a substrate for signal peptide peptidase-like 3 (SPPL3).J Biol Chem. 2012 Dec 21;287(52):43401-9. doi: 10.1074/jbc.M112.371369. Epub 2012 Nov 6. J Biol Chem. 2012. PMID: 23132852 Free PMC article.
-
Viruses and Metabolism: The Effects of Viral Infections and Viral Insulins on Host Metabolism.Annu Rev Virol. 2021 Sep 29;8(1):373-391. doi: 10.1146/annurev-virology-091919-102416. Annu Rev Virol. 2021. PMID: 34586876 Free PMC article. Review.
-
Sll0528, a Site-2-Protease, Is Critically Involved in Cold, Salt and Hyperosmotic Stress Acclimation of Cyanobacterium Synechocystis sp. PCC 6803.Int J Mol Sci. 2014 Dec 8;15(12):22678-22693. doi: 10.3390/ijms151222678. Int J Mol Sci. 2014. PMID: 25493476 Free PMC article.
-
Involvement of a Membrane-Bound Amphiphilic Helix in Substrate Discrimination and Binding by an Escherichia coli S2P Peptidase RseP.Front Microbiol. 2020 Nov 27;11:607381. doi: 10.3389/fmicb.2020.607381. eCollection 2020. Front Microbiol. 2020. PMID: 33329500 Free PMC article.
-
Metallopeptidase Stp1 activates the transcription factor Sre1 in the carotenogenic yeast Xanthophyllomyces dendrorhous.J Lipid Res. 2020 Feb;61(2):229-243. doi: 10.1194/jlr.RA119000431. Epub 2019 Dec 5. J Lipid Res. 2020. PMID: 31806730 Free PMC article.
References
-
- Freeman M. Rhomboid proteases and their biological functions. Ann Rev Gen. 2008;42:191–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

