Horizontal gene transfer and the genomics of enterococcal antibiotic resistance
- PMID: 20837397
- PMCID: PMC2955785
- DOI: 10.1016/j.mib.2010.08.004
Horizontal gene transfer and the genomics of enterococcal antibiotic resistance
Abstract
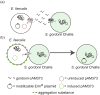
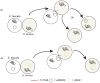
Enterococci are Gram-positive bacteria that normally colonize gastrointestinal tracts of humans and animals. They are of growing concern because of their ability to cause antibiotic resistant hospital infections. Antibiotic resistance has been acquired, and has disseminated throughout enterococci, via horizontal transfer of mobile genetic elements. This transmission has been mediated mainly by conjugative plasmids of the pheromone-responsive and broad host range incompatibility group 18 type. Genome sequencing is revealing the extent of diversity of these and other mobile elements in enterococci, as well as the extent of recombination and rearrangement resulting in new phenotypes. Pheromone-responsive plasmids were recently shown to promote genome plasticity in antibiotic resistant Enterococcus faecalis, and their involvement has been implicated in E. faecium as well. Further, incompatibility group 18 plasmids have recently played an important role in mediating transfer of vancomycin resistance from enterococci to methicillin-resistant strains of S. aureus.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. - PubMed
-
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

