G-helix of maspin mediates effects on cell migration and adhesion
- PMID: 20837467
- PMCID: PMC2978556
- DOI: 10.1074/jbc.M110.177253
G-helix of maspin mediates effects on cell migration and adhesion
Abstract
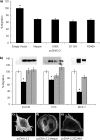
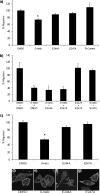
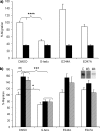
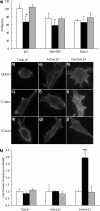
Maspin is a member of the serine protease inhibitor (serpin) superfamily that lacks protease inhibitory ability, although displaying tumor metastasis-suppressing activity resulting from its influence on cell migration, invasion, proliferation, apoptosis, and adhesion. The molecular mechanisms of these actions of maspin are as yet undefined. Here, we sought to identify critical functional motifs by the expression of maspin with point mutations at sites potentially involved in protein-protein interactions: the G α-helix (G-helix), an internal salt bridge or the P1 position of the reactive center loop. Our findings indicate that only mutations in the G-helix attenuated inhibition of cell migration by maspin and that this structural element is also involved in the effect of maspin on cell adhesion. The action of maspin on cell migration could be mimicked by a 15-mer G-helix peptide, indicating that the G-helix is both essential and sufficient for this effect. In addition, we provide evidence that the effects of the G-helix of maspin are dependent on β1 integrins. These data reveal that the major extracellular functions associated with the tumor suppressive action of maspin likely involve interactions in which the G-helix plays a key role.
Figures

Similar articles
-
Maspin functions as tumor suppressor by increasing cell adhesion to extracellular matrix in prostate tumor cells.J Urol. 2003 Mar;169(3):1157-61. doi: 10.1097/01.ju.0000040245.70349.37. J Urol. 2003. PMID: 12576872
-
Maspin is physically associated with [beta]1 integrin regulating cell adhesion in mammary epithelial cells.FASEB J. 2006 Jul;20(9):1510-2. doi: 10.1096/fj.05-5500fje. Epub 2006 May 23. FASEB J. 2006. PMID: 16720730
-
Identification of novel peptide motifs in the serpin maspin that affect vascular smooth muscle cell function.Biochim Biophys Acta Mol Cell Res. 2017 Feb;1864(2):336-344. doi: 10.1016/j.bbamcr.2016.11.019. Epub 2016 Nov 23. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 27888098
-
Maspin--a novel protease inhibitor with tumor-suppressing activity in breast cancer.Acta Oncol. 2000;39(8):931-4. doi: 10.1080/02841860050215909. Acta Oncol. 2000. PMID: 11206999 Review.
-
Maspin. A tumor suppressing serpin.Adv Exp Med Biol. 1997;425:77-88. Adv Exp Med Biol. 1997. PMID: 9433491 Review.
Cited by
-
Decreased maspin combined with elevated vascular endothelial growth factor C is associated with poor prognosis in non-small cell lung cancer.Thorac Cancer. 2014 Sep;5(5):383-90. doi: 10.1111/1759-7714.12104. Epub 2014 Aug 25. Thorac Cancer. 2014. PMID: 26767029 Free PMC article.
-
SERPINB5 Expression: Association with CCRT Response and Prognostic Value in Rectal Cancer.Int J Med Sci. 2018 Feb 12;15(4):376-384. doi: 10.7150/ijms.22823. eCollection 2018. Int J Med Sci. 2018. PMID: 29511373 Free PMC article.
-
Supramolecular assembly of multifunctional maspin-mimetic nanostructures as a potent peptide-based angiogenesis inhibitor.Acta Biomater. 2015 Jan;12:1-10. doi: 10.1016/j.actbio.2014.11.001. Epub 2014 Nov 8. Acta Biomater. 2015. PMID: 25462852 Free PMC article.
-
The Opportunity of Precision Medicine for Breast Cancer With Context-Sensitive Tumor Suppressor Maspin.J Cell Biochem. 2017 Jul;118(7):1639-1647. doi: 10.1002/jcb.25969. Epub 2017 Mar 21. J Cell Biochem. 2017. PMID: 28262971 Free PMC article. Review.
-
Maspin binds to cardiolipin in mitochondria and triggers apoptosis.FASEB J. 2019 May;33(5):6354-6364. doi: 10.1096/fj.201802182R. Epub 2019 Feb 20. FASEB J. 2019. PMID: 30786218 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

