O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids
- PMID: 20837471
- PMCID: PMC2978557
- DOI: 10.1074/jbc.M110.156950
O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids
Abstract
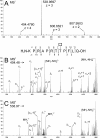
The angiopoietin-like protein 3 (ANGPTL3) is an important inhibitor of the endothelial and lipoprotein lipases and a promising drug target. ANGPTL3 undergoes proprotein convertase processing (RAPR(224)↓TT) for activation, and the processing site contains two potential GalNAc O-glycosylation sites immediately C-terminal (TT(226)). We developed an in vivo model system in CHO ldlD cells that was used to show that O-glycosylation in the processing site blocked processing of ANGPTL3. Genome-wide SNP association studies have identified the polypeptide GalNAc-transferase gene, GALNT2, as a candidate gene for low HDL and high triglyceride blood levels. We hypothesized that the GalNAc-T2 transferase performed critical O-glycosylation of proteins involved in lipid metabolism. Screening of a panel of proteins known to affect lipid metabolism for potential sites glycosylated by GalNAc-T2 led to identification of Thr(226) adjacent to the proprotein convertase processing site in ANGPTL3. We demonstrated that GalNAc-T2 glycosylation of Thr(226) in a peptide with the RAPR(224)↓TT processing site blocks in vitro furin cleavage. The study demonstrates that ANGPTL3 activation is modulated by O-glycosylation and that this step is probably controlled by GalNAc-T2.
Figures





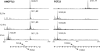

References
-
- Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., Clausen H. (2006) J. Biol. Chem. 281, 18370–18377 - PubMed
-
- Steiner D. F. (1998) Curr. Opin. Chem. Biol. 2, 31–39 - PubMed
-
- Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., Sprecher E. (2004) Nat. Genet. 36, 579–581 - PubMed
-
- Benet-Pagès A., Orlik P., Strom T. M., Lorenz-Depiereux B. (2005) Hum. Mol. Genet. 14, 385–390 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

