Differential roles of Sall4 isoforms in embryonic stem cell pluripotency
- PMID: 20837710
- PMCID: PMC2976381
- DOI: 10.1128/MCB.00419-10
Differential roles of Sall4 isoforms in embryonic stem cell pluripotency
Abstract
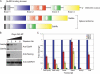
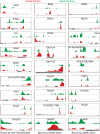
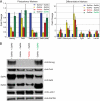
Murine embryonic stem (ES) cells are defined by continuous self-renewal and pluripotency. A diverse repertoire of protein isoforms arising from alternative splicing is expressed in ES cells without defined biological roles. Sall4, a transcription factor essential for pluripotency, exists as two isoforms (Sall4a and Sall4b). Both isoforms can form homodimers and a heterodimer with each other, and each can interact with Nanog. By genomewide location analysis, we determined that Sall4a and Sall4b have overlapping, but not identical binding sites within the ES cell genome. In addition, Sall4b, but not Sall4a, binds preferentially to highly expressed loci in ES cells. Sall4a and Sall4b binding sites are distinguished by both epigenetic marks at target loci and their clustering with binding sites of other pluripotency factors. When ES cells expressing a single isoform of Sall4 are generated, Sall4b alone could maintain the pluripotent state, although it could not completely suppress all differentiation markers. Sall4a and Sall4b collaborate in maintenance of the pluripotent state but play distinct roles. Our work is novel in establishing such isoform-specific differences in ES cells.
Figures










References
-
- Al-Baradie, R., K. Yamada, C. St. Hilaire, W. M. Chan, C. Andrews, N. McIntosh, M. Nakano, E. J. Martonyi, W. R. Raymond, S. Okumura, M. M. Okihiro, and E. C. Engle. 2002. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am. J. Hum. Genet. 71:1195-1199. - PMC - PubMed
-
- Atlasi, Y., S. J. Mowla, S. A. Ziaee, P. J. Gokhale, and P. W. Andrews. 2008. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells 26:3068-3074. - PubMed
-
- Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315-326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
