Reversal of salt preference is directed by the insulin/PI3K and Gq/PKC signaling in Caenorhabditis elegans
- PMID: 20837997
- PMCID: PMC2998313
- DOI: 10.1534/genetics.110.119768
Reversal of salt preference is directed by the insulin/PI3K and Gq/PKC signaling in Caenorhabditis elegans
Abstract
Animals search for foods and decide their behaviors according to previous experience. Caenorhabditis elegans detects chemicals with a limited number of sensory neurons, allowing us to dissect roles of each neuron for innate and learned behaviors. C. elegans is attracted to salt after exposure to the salt (NaCl) with food. In contrast, it learns to avoid the salt after exposure to the salt without food. In salt-attraction behavior, it is known that the ASE taste sensory neurons (ASEL and ASER) play a major role. However, little is known about mechanisms for learned salt avoidance. Here, through dissecting contributions of ASE neurons for salt chemotaxis, we show that both ASEL and ASER generate salt chemotaxis plasticity. In ASER, we have previously shown that the insulin/PI 3-kinase signaling acts for starvation-induced salt chemotaxis plasticity. This study shows that the PI 3-kinase signaling promotes aversive drive of ASER but not of ASEL. Furthermore, the Gq signaling pathway composed of Gqα EGL-30, diacylglycerol, and nPKC (novel protein kinase C) TTX-4 promotes attractive drive of ASER but not of ASEL. A putative salt receptor GCY-22 guanylyl cyclase is required in ASER for both salt attraction and avoidance. Our results suggest that ASEL and ASER use distinct molecular mechanisms to regulate salt chemotaxis plasticity.
Figures
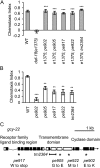
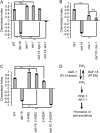

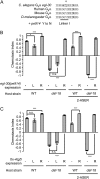


References
-
- Bargmann, C., 2006. Chemosensation in C. elegans (October 25, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.123.1, http://www.wormbook.org.
-
- Bargmann, C., and H. Horvitz, 1991. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7 729–742. - PubMed
-
- Bargmann, C., E. Hartwieg and H. Horvitz, 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74 515–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

