Preferential re-replication of Drosophila heterochromatin in the absence of geminin
- PMID: 20838463
- PMCID: PMC2936543
- DOI: 10.1371/journal.pgen.1001112
Preferential re-replication of Drosophila heterochromatin in the absence of geminin
Abstract
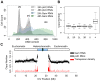
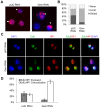
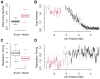
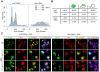
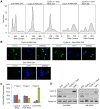
To ensure genomic integrity, the genome must be duplicated exactly once per cell cycle. Disruption of replication licensing mechanisms may lead to re-replication and genomic instability. Cdt1, also known as Double-parked (Dup) in Drosophila, is a key regulator of the assembly of the pre-replicative complex (pre-RC) and its activity is strictly limited to G1 by multiple mechanisms including Cul4-Ddb1 mediated proteolysis and inhibition by geminin. We assayed the genomic consequences of disregulating the replication licensing mechanisms by RNAi depletion of geminin. We found that not all origins of replication were sensitive to geminin depletion and that heterochromatic sequences were preferentially re-replicated in the absence of licensing mechanisms. The preferential re-activation of heterochromatic origins of replication was unexpected because these are typically the last sequences to be duplicated in a normal cell cycle. We found that the re-replication of heterochromatin was regulated not at the level of pre-RC activation, but rather by the formation of the pre-RC. Unlike the global assembly of the pre-RC that occurs throughout the genome in G1, in the absence of geminin, limited pre-RC assembly was restricted to the heterochromatin by elevated cyclin A-CDK activity. These results suggest that there are chromatin and cell cycle specific controls that regulate the re-assembly of the pre-RC outside of G1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
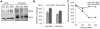
References
-
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14(18):778–786. - PubMed
-
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. - PubMed
-
- Nguyen V, Co C, Irie K, Li J. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10:195–205. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

