Evidence that mutation is universally biased towards AT in bacteria
- PMID: 20838599
- PMCID: PMC2936535
- DOI: 10.1371/journal.pgen.1001115
Evidence that mutation is universally biased towards AT in bacteria
Abstract
Mutation is the engine that drives evolution and adaptation forward in that it generates the variation on which natural selection acts. Mutation is a random process that nevertheless occurs according to certain biases. Elucidating mutational biases and the way they vary across species and within genomes is crucial to understanding evolution and adaptation. Here we demonstrate that clonal pathogens that evolve under severely relaxed selection are uniquely suitable for studying mutational biases in bacteria. We estimate mutational patterns using sequence datasets from five such clonal pathogens belonging to four diverse bacterial clades that span most of the range of genomic nucleotide content. We demonstrate that across different types of sites and in all four clades mutation is consistently biased towards AT. This is true even in clades that have high genomic GC content. In all studied cases the mutational bias towards AT is primarily due to the high rate of C/G to T/A transitions. These results suggest that bacterial mutational biases are far less variable than previously thought. They further demonstrate that variation in nucleotide content cannot stem entirely from variation in mutational biases and that natural selection and/or a natural selection-like process such as biased gene conversion strongly affect nucleotide content.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
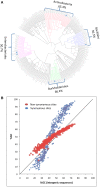
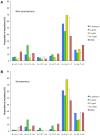
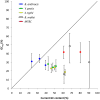
 . GCeq values are significantly lower than the observed GC content at all site categories (intergenic, synonymous and non-synonymous) and for all four lineages of clonal pathogens with either intermediate or high GC contents. MTBC intergenic SNPs were not available for analysis. Error bars depict 95% confidence intervals for GCeq. No correlation is observed between GCeq and current GC for the clonal pathogen lineages (P≤1).
. GCeq values are significantly lower than the observed GC content at all site categories (intergenic, synonymous and non-synonymous) and for all four lineages of clonal pathogens with either intermediate or high GC contents. MTBC intergenic SNPs were not available for analysis. Error bars depict 95% confidence intervals for GCeq. No correlation is observed between GCeq and current GC for the clonal pathogen lineages (P≤1).
References
-
- Lynch M. The origins of Genome Architecture. Sunderland, MA: Sinauer Associates, Inc; 2007.
-
- Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

