Computational systems chemical biology
- PMID: 20838980
- PMCID: PMC3547368
- DOI: 10.1007/978-1-60761-839-3_18
Computational systems chemical biology
Abstract
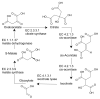
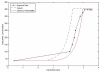
There is a critical need for improving the level of chemistry awareness in systems biology. The data and information related to modulation of genes and proteins by small molecules continue to accumulate at the same time as simulation tools in systems biology and whole body physiologically based pharmacokinetics (PBPK) continue to evolve. We called this emerging area at the interface between chemical biology and systems biology systems chemical biology (SCB) (Nat Chem Biol 3: 447-450, 2007).The overarching goal of computational SCB is to develop tools for integrated chemical-biological data acquisition, filtering and processing, by taking into account relevant information related to interactions between proteins and small molecules, possible metabolic transformations of small molecules, as well as associated information related to genes, networks, small molecules, and, where applicable, mutants and variants of those proteins. There is yet an unmet need to develop an integrated in silico pharmacology/systems biology continuum that embeds drug-target-clinical outcome (DTCO) triplets, a capability that is vital to the future of chemical biology, pharmacology, and systems biology. Through the development of the SCB approach, scientists will be able to start addressing, in an integrated simulation environment, questions that make the best use of our ever-growing chemical and biological data repositories at the system-wide level. This chapter reviews some of the major research concepts and describes key components that constitute the emerging area of computational systems chemical biology.
Figures






References
4. References
-
- Amidon GL, Lennernäs H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutics drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm. Res. 1995;12:413–420. - PubMed
-
- Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. - PubMed
-
- Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. - PubMed
-
- Biocarta is a commercially-sponsored “open source” forum that integrates emerging proteomic information from the scientific community and depicts inter-molecular interactions via dynamic graphical models. http://www.biocarta.com/genes/index.asp.
Reference List
-
- FDA Approved Drug Products. FDA 2009a. Ref Type: Electronic Citation.
-
- EGG 2009b http://www.genome.jp/kegg/ Ref Type: Electronic Citation.
-
- Physicians’ Desk Reference. PDR. 2009c.
-
- SciFinder: American Chemical Society CAS online / SciFinder. 2009d http://www.cas.org/SCIFINDER/ Ref Type: Electronic Citation.
-
- Amidon GL, Lennernas H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm Res. 1995;12:413–420. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

