Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes
- PMID: 20841432
- PMCID: PMC2963823
- DOI: 10.1101/gr.108332.110
Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes
Abstract
The human gut microbiome is a complex ecosystem composed mainly of uncultured bacteria. It plays an essential role in the catabolism of dietary fibers, the part of plant material in our diet that is not metabolized in the upper digestive tract, because the human genome does not encode adequate carbohydrate active enzymes (CAZymes). We describe a multi-step functionally based approach to guide the in-depth pyrosequencing of specific regions of the human gut metagenome encoding the CAZymes involved in dietary fiber breakdown. High-throughput functional screens were first applied to a library covering 5.4 × 10(9) bp of metagenomic DNA, allowing the isolation of 310 clones showing beta-glucanase, hemicellulase, galactanase, amylase, or pectinase activities. Based on the results of refined secondary screens, sequencing efforts were reduced to 0.84 Mb of nonredundant metagenomic DNA, corresponding to 26 clones that were particularly efficient for the degradation of raw plant polysaccharides. Seventy-three CAZymes from 35 different families were discovered. This corresponds to a fivefold target-gene enrichment compared to random sequencing of the human gut metagenome. Thirty-three of these CAZy encoding genes are highly homologous to prevalent genes found in the gut microbiome of at least 20 individuals for whose metagenomic data are available. Moreover, 18 multigenic clusters encoding complementary enzyme activities for plant cell wall degradation were also identified. Gene taxonomic assignment is consistent with horizontal gene transfer events in dominant gut species and provides new insights into the human gut functional trophic chain.
Figures
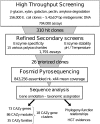
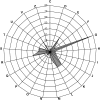
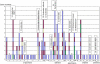

References
-
- Chessel D, Dufour AB, Thioulouse J 2004. The ade4 package—I: One-table methods. R News 4: 5–10
-
- Duan CJ, Xian L, Zhao GC, Feng Y, Pang H, Bai XL, Tang JL, Ma QS, Feng JX 2009. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol 107: 245–256 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
