Small self-cleaving ribozymes
- PMID: 20843979
- PMCID: PMC2944367
- DOI: 10.1101/cshperspect.a003574
Small self-cleaving ribozymes
Abstract
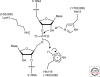
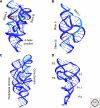
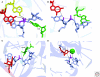
The hammerhead, hairpin, hepatitis delta virus (HDV), Varkud Satellite (VS), and glmS ribozymes catalyze sequence-specific intramolecular cleavage of RNA. They range between 50 and 150 nucleotides in length, and are known as the "small self-cleaving ribozymes." Except for the glmS ribozyme that functions as a riboswitch in Gram-positive bacteria, they were originally discovered as domains of satellite RNAs. However, recent studies show that several of them are broadly distributed in genomes of organisms from many phyla. Each of these ribozymes has a unique overall architecture and active site organization. Crystal structures have revealed how RNA active sites can bind preferentially to the transition state of a reaction, whereas mechanistic studies have shown that nucleobases can efficiently perform general acid-base and electrostatic catalysis. This versatility explains the abundance of ribozymes in contemporary organisms and also supports a role for catalytic RNAs early in evolution.
Figures
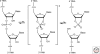
References
-
- Bevilacqua PC 2003. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry 42: 2259–2265 - PubMed
-
- Bevilacqua PC, Yajima R 2006. Nucleobase catalysis in ribozyme mechanism. Current Op Chem Biol 10: 455–464 - PubMed
-
- Blount KF, Uhlenbeck OC 2002. Internal equilibrium of the hammerhead ribozyme is altered by the length of certain covalent cross-links. Biochemistry 41: 6834–6841 - PubMed
-
- Blount KF, Uhlenbeck OC 2005. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct 34: 410–440 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
