sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments
- PMID: 20843985
- PMCID: PMC2957045
- DOI: 10.1261/rna.2193110
sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments
Abstract
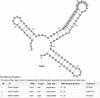
Bacterial sRNAs are an emerging class of small regulatory RNAs, 40-500 nt in length, which play a variety of important roles in many biological processes through binding to their mRNA or protein targets. A comprehensive database of experimentally confirmed sRNA targets would be helpful in understanding sRNA functions systematically and provide support for developing prediction models. Here we report on such a database--sRNATarBase. The database holds 138 sRNA-target interactions and 252 noninteraction entries, which were manually collected from peer-reviewed papers. The detailed information for each entry, such as supporting experimental protocols, BLAST-based phylogenetic analysis of sRNA-mRNA target interaction in closely related bacteria, predicted secondary structures for both sRNAs and their targets, and available binding regions, is provided as accurately as possible. This database also provides hyperlinks to other databases including GenBank, SWISS-PROT, and MPIDB. The database is available from the web page http://ccb.bmi.ac.cn/srnatarbase/.
Figures



References
-
- Backofen R, Hess WR 2010. Computational prediction of sRNAs and their targets in bacteria. RNA Biol 7: 1–10 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
