Interactions between Hedgehog proteins and their binding partners come into view
- PMID: 20844013
- PMCID: PMC2939362
- DOI: 10.1101/gad.1951710
Interactions between Hedgehog proteins and their binding partners come into view
Abstract
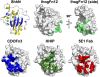
Hedgehog (Hh) proteins are secreted signaling molecules that mediate essential tissue-patterning events during embryonic development and function in tissue homeostasis and regeneration throughout life. Hh signaling is regulated by multiple mechanisms, including covalent lipid modification of the Hh protein and interactions with multiple protein and glycan partners. Unraveling the nature and effects of these interactions has proven challenging, but recent structural and biophysical studies of Hh proteins and active fragments of heparin, Ihog, Cdo, Boc, Hedgehog-interacting protein (Hhip), Patched (Ptc), and the monoclonal antibody 5E1 have added a new level of molecular detail to our understanding of how Hh signal response and distribution are regulated within tissues. We review these results and discuss their implications for understanding Hh signaling in normal and disease states.
Figures

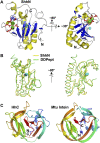
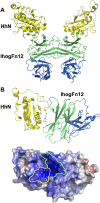
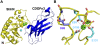


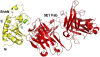
References
-
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE 1996. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86: 221–232 - PubMed
-
- Amanai K, Jiang J 2001. Distinct roles of Central missing and Dispatched in sending the Hedgehog signal. Development 128: 5119–5127 - PubMed
-
- Beckett K, Franch-Marro X, Vincent JP 2008. Glypican-mediated endocytosis of Hedgehog has opposite effects in flies and mice. Trends Cell Biol 18: 360–363 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
