Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries
- PMID: 20844017
- PMCID: PMC2939368
- DOI: 10.1101/gad.1963210
Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries
Abstract
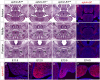
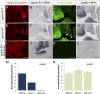
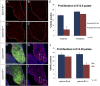
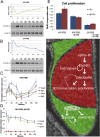
Mutations in the X-linked human EPHRIN-B1 gene result in cleft palate and other craniofacial anomalies as part of craniofrontonasal syndrome (CFNS), but the molecular and developmental mechanisms by which ephrin-B1 controls the underlying developmental processes are not clear. Here we demonstrate that ephrin-B1 plays an intrinsic role in palatal shelf outgrowth in the mouse by regulating cell proliferation in the anterior palatal shelf mesenchyme. In ephrin-B1 heterozygous mutants, X inactivation generates ephrin-B1-expressing and -nonexpressing cells that sort out, resulting in mosaic ephrin-B1 expression. We now show that this process leads to mosaic disruption of cell proliferation and post-transcriptional up-regulation of EphB receptor expression through relief of endocytosis and degradation. The alteration in proliferation rates resulting from ectopic Eph-ephrin expression boundaries correlates with the more severe dysmorphogenesis of ephrin-B1(+/-) heterozygotes that is a hallmark of CFNS. Finally, by integrating phosphoproteomic and transcriptomic approaches, we show that ephrin-B1 controls proliferation in the palate by regulating the extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) signal transduction pathway.
Figures



References
-
- Bentires-Alj M, Kontaridis MI, Neel BG 2006. Stops along the RAS pathway in human genetic disease. Nat Med 12: 283–285 - PubMed
-
- Braybrook C, Lisgo S, Doudney K, Henderson D, Marcano AC, Strachan T, Patton MA, Villard L, Moore GE, Stanier P, et al. 2002. Craniofacial expression of human and murine TBX22 correlates with the cleft palate and ankyloglossia phenotype observed in CPX patients. Hum Mol Genet 11: 2793–2804 - PubMed
-
- Bush JO, Lan Y, Maltby KM, Jiang R 2002. Isolation and developmental expression analysis of Tbx22, the mouse homolog of the human X-linked cleft palate gene. Dev Dyn 225: 322–326 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
