The varicella-zoster virus ORFS/L (ORF0) gene is required for efficient viral replication and contains an element involved in DNA cleavage
- PMID: 20844039
- PMCID: PMC2977885
- DOI: 10.1128/JVI.00878-10
The varicella-zoster virus ORFS/L (ORF0) gene is required for efficient viral replication and contains an element involved in DNA cleavage
Abstract
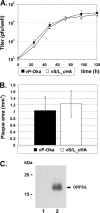
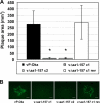
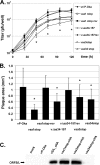
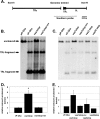
The genome of varicella-zoster virus (VZV), a human alphaherpesvirus, consists of two unique regions, unique long (U(L)) and unique short (U(S)), each of which is flanked by inverted repeats. During replication, four isomers of the viral DNA are generated which are distinguished by the relative orientations of U(L) and U(S). VZV virions predominantly package two isomeric forms of the genome that have a fixed orientation of U(L). An open reading frame (ORF) of unknown function, ORFS/L, also referred to as ORF0, is located at the extreme terminus of U(L), directly adjacent to the a-like sequences, which are known to be involved in cleavage and packaging of viral DNA. We demonstrate here that the ORFS/L protein localizes to the Golgi network in infected and transfected cells. Furthermore, we were able to demonstrate that deletion of the predicted ORFS/L gene is lethal, while retention of the N-terminal 28 amino acid residues resulted in viable yet replication-impaired virus. The growth defect was only partially attributable to the expression of the ORFS/L product, suggesting that the 5' region of ORFS/L contains a sequence element crucial for cleavage/packaging of viral DNA. Consequently, mutations introduced into the extreme 5' terminus of ORFS/L resulted in a defect in DNA cleavage, indicating that the region is indeed involved in the processing of viral DNA. Since the sequence element has no counterpart at the other end of U(L), we concluded that our results can provide an explanation for the almost exclusive orientation of the U(L) seen in packaged VZV DNA.
Figures


Similar articles
-
Sequence analysis of the leftward end of simian varicella virus (EcoRI-I fragment) reveals the presence of an 8-bp repeat flanking the unique long segment and an 881-bp open-reading frame that is absent in the varicella zoster virus genome.Virology. 2000 Sep 1;274(2):420-8. doi: 10.1006/viro.2000.0465. Virology. 2000. PMID: 10964784
-
The varicella-zoster virus portal protein is essential for cleavage and packaging of viral DNA.J Virol. 2014 Jul;88(14):7973-86. doi: 10.1128/JVI.00376-14. Epub 2014 May 7. J Virol. 2014. PMID: 24807720 Free PMC article.
-
Synthesis and decay of varicella zoster virus transcripts.J Neurovirol. 2011 Jun;17(3):281-7. doi: 10.1007/s13365-011-0029-2. Epub 2011 Apr 12. J Neurovirol. 2011. PMID: 21484478 Free PMC article.
-
Varicella zoster virus: out of Africa and into the research laboratory.Herpes. 2006 Aug;13(2):32-6. Herpes. 2006. PMID: 16895651 Review.
-
Lessons to be learned from varicella-zoster virus.Vet Microbiol. 1996 Nov;53(1-2):55-66. doi: 10.1016/s0378-1135(96)01234-5. Vet Microbiol. 1996. PMID: 9010998 Review.
Cited by
-
Comparative Analysis of the Simian Varicella Virus and Varicella Zoster Virus Genomes.Viruses. 2022 Apr 19;14(5):844. doi: 10.3390/v14050844. Viruses. 2022. PMID: 35632586 Free PMC article. Review.
-
Molecular Aspects of Varicella-Zoster Virus Latency.Viruses. 2018 Jun 28;10(7):349. doi: 10.3390/v10070349. Viruses. 2018. PMID: 29958408 Free PMC article. Review.
-
Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation.J Exp Med. 2011 Mar 14;208(3):605-15. doi: 10.1084/jem.20101402. Epub 2011 Mar 7. J Exp Med. 2011. PMID: 21383055 Free PMC article.
-
Creating the "Dew Drop on a Rose Petal": the Molecular Pathogenesis of Varicella-Zoster Virus Skin Lesions.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0011622. doi: 10.1128/mmbr.00116-22. Epub 2023 Jun 24. Microbiol Mol Biol Rev. 2023. PMID: 37354037 Free PMC article. Review.
-
Simian varicella virus gene expression during acute and latent infection of rhesus macaques.J Neurovirol. 2011 Dec;17(6):600-12. doi: 10.1007/s13365-011-0057-y. Epub 2011 Nov 4. J Neurovirol. 2011. PMID: 22052378 Free PMC article.
References
-
- Arvin, A. M. 2001. Varicella-zoster virus, p. 2731-2767. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lippincott-Raven Publishers, Philadelphia, PA.
-
- Berkowitz, C., M. Moyal, A. Rosen-Wolff, G. Darai, and Y. Becker. 1994. Herpes simplex virus type 1 (HSV-1) UL56 gene is involved in viral intraperitoneal pathogenicity to immunocompetent mice. Arch. Virol. 134:73-83. - PubMed
-
- Bruce, M. C., V. Kanelis, F. Fouladkou, A. Debonneville, O. Staub, and D. Rotin. 2008. Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem. J. 415:155-163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

